Use of pgSIT technology on the Onetahi island of the Tetiaroa atoll in French Polynesia.
Data sources and sims:
- MGDrivE sim (scripts to generate datasets)
- Transforming insect population control with precision guided sterile males with demonstration in flies
- Reply to ‘Concerns about the feasibility of using “precision guided sterile males” to control insects’
Folders and files follow this naming convention:
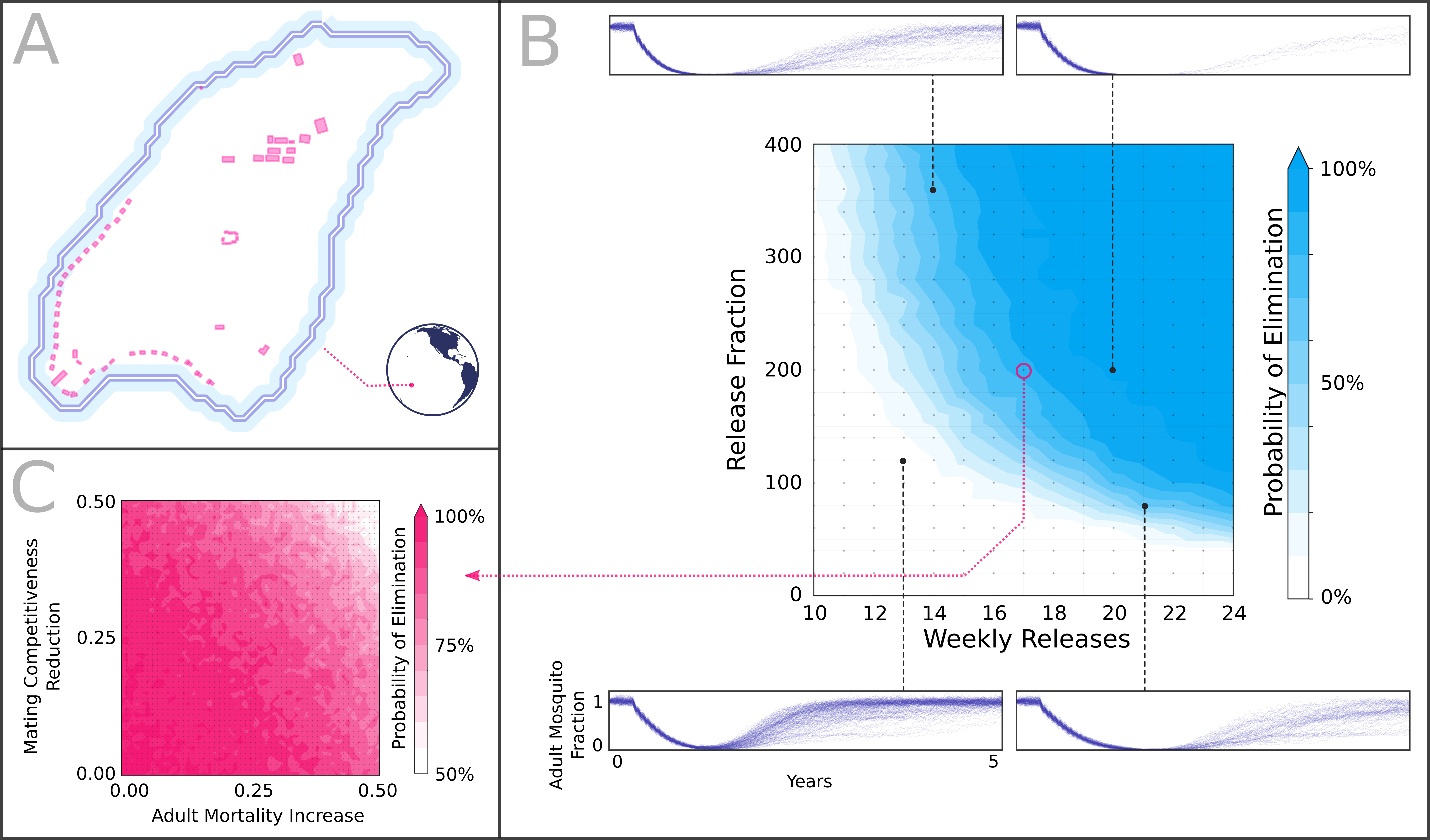
E_pop_ren_res_mad_matpop: Population size (male and female) per node (x1)ren: Number of weekly releases (x1)res: Release size (fraction of the stable population x100)mad: Adult lifespan reduction (x100)mat: Male mating reduction (x100)
For the breakdown of the AOI sets, look at the pgSIT gene definition.
Exported metrics (MOI) for the drive are:
- POE: Probability of elimination
- WOP: Total sum of time below the threshold
- TTI: First break below the threshold
- TTO: Last break below the threshold
- RAP: Fraction of the population with the genes at given points of time
- MNX: Minimum and maximum of genes in the population
Summary statistic files follow this naming convention:
AOI_MOI_QNT_qnt.csv
Where the main AOI was HLT (presence of mosquitoes) and the outputs (labels) are:
- TTI, TTO, WOP: Fraction's threshold for the metric to be true
- RAP: Fraction of present genotypes at given points (days) of the simulation
- POE: Probability of eliminating mosquitoes given the stochastic runs
- MNX: Min/Max and days at which these are achieved
./PYF_preProcess.sh USR LND PLOTS_BOOL./PYF_pstProcess.sh USR LND PLOTS_BOOL HEAT_BOOL./PYF_clsPipeline.sh USR LND
After running the ML pipeline, load the dashboard with:
python PYF_clsApp.py PATH_TO_MODELS
Go to http://127.0.0.1:8050/