📈 Examine the impact of COVID-19 spread by using a custom epidemiological model.
Simulation and modeling is becoming a standard approach to understand complex bio- chemical processes. Therefore, there is a big need for software tools that allow access to diverse simulation and modeling methods as well as support for the usage of these method. COPASI is an open-source software application for creating and solving mathematical models of biological processes such as metabolic networks, cell-signaling pathways, reg- ulatory networks, infectious diseases, and many others. In this case, this software was utilized as an epidemic simulator since the the biochemical reactions networks resemble the viruses behaviour.
As an extra step, the same model without the vaccination part, was implemented in python (see SIERD_model) for this project as well.
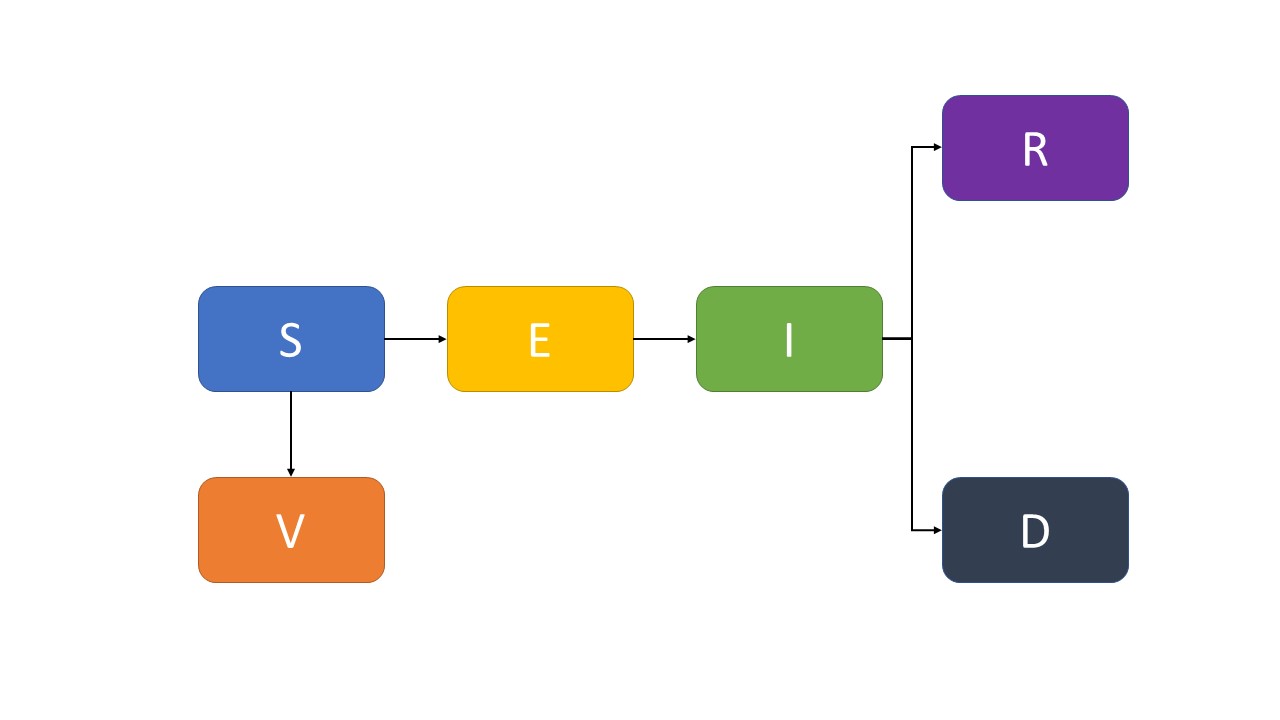
The basic SEIR model is expanded to six compartments to simulate the epidemic of COVID-19. Six state variables are considered within a population, that is, S(t), E(t), I(t), R(t), D(t), and V(t), denoting the number of susceptible, exposed (infected, but not yet infectious), infectious (symptomatic and asymptomatic), recovered, dead, and vacci- nated cases, respectively. Thus, the human population is denoted by N(t) = S(t)+E(t)+ I(t) + R(t) + D(t) + V (t).
The model culminates to a six-dimensional system of ordinary differential equations as follows.
The parameters used in the COVID-19 transmission model di�rentials equations are given in the following table.
| Model parameter name | Meaning | Case/Reference |
|---|---|---|
| alpha | Vaccination rate | 0.04 per day [1] |
| beta | Contact rate | ASSUMED |
| gamma | Mean recovery rate | 0.06 per day [2] |
| sigma | Incubation rate | 0.09 [2] |
| p_d | Covid Fatality rate | 0.0018 [2] |
-
Import SIERD-V.cps into COPASI
-
To run the model for the initial parameters, go to 'Tasks>Time Course', create a plot by pressing the button 'Output Assistant' and hit 'Run'.
-
To apply Sensitivity Analysis, head over to 'Tasks > Parameter Scan', press 'Create' and choose a global quantity. Then, you are ready to to apply sensitivity analysis by pressing 'Run'.
Change parameters to your liking
[1] E. O.-O. Max Roser, Hannah Ritchie and J. Hasell. Coronavirus pandemic (covid-19).
Our World in Data, 2020. https://ourworldindata.org/coronavirus.
[2] S. Mwalili, M. Kimathi, V. Ojiambo, D. Gathungu, and R. Mbogo. SEIR model
for COVID-19 dynamics incorporating the environment and social distancing. BMC
Research Notes, 13(1), July 2020.
[3] W. C. Roda, M. B. Varughese, D. Han, and M. Y. Li. Why is it diffcult to accurately
predict the COVID-19 epidemic? Infectious Disease Modelling, 5:271{281, 2020.