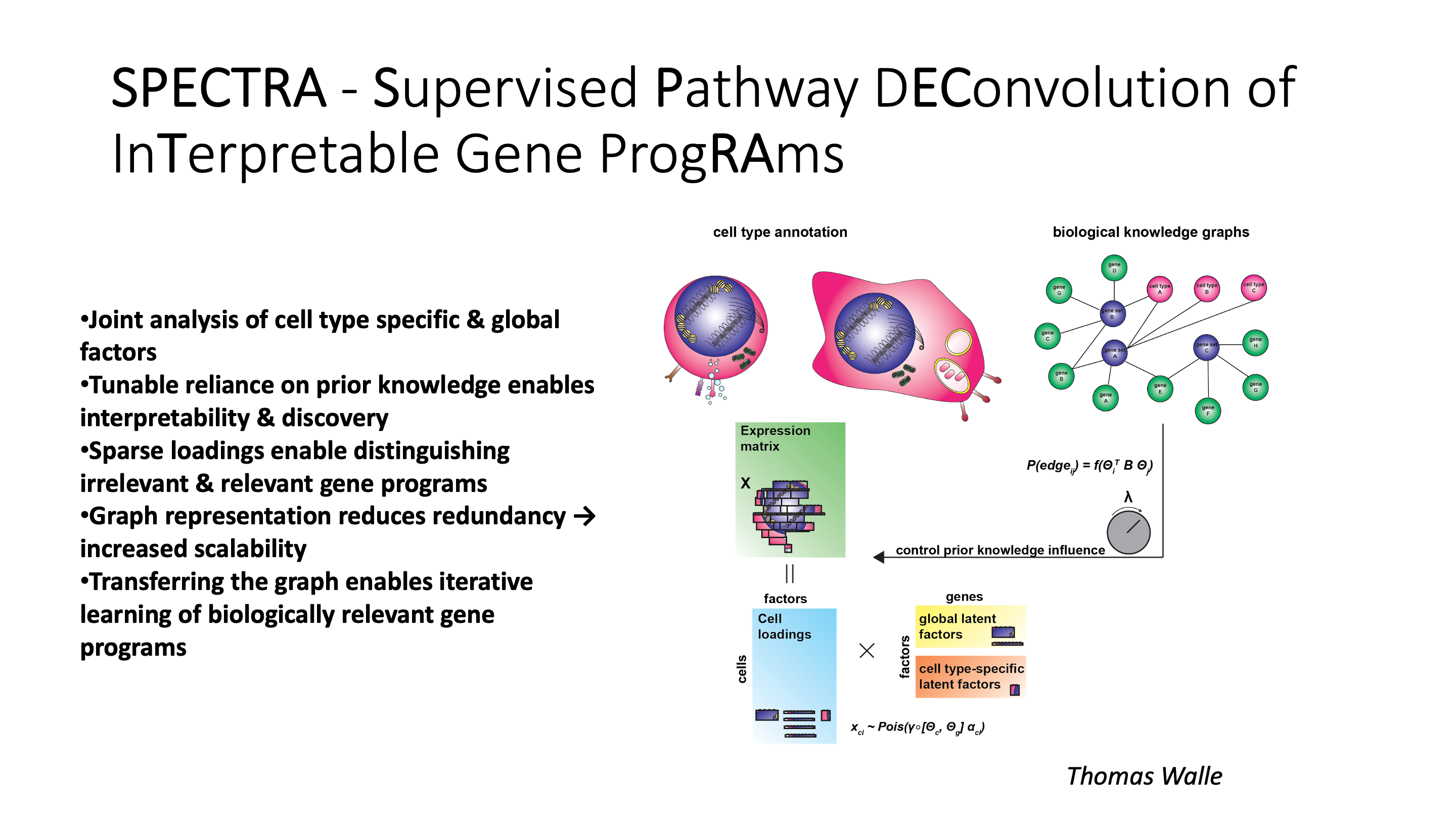
SPECTRA takes in a single cell gene expression matrix and a set of pathway annotations to fit the data to.
gene_set_annotations = {
"global": {"global_ifn_II_response" : ["CIITA", "CXCL10", ...] , "global_MHCI": ["HLA-A". "HLA-B", "HLA-C"] },
"CD8_T": {"CD8_exhaustion" : ["CXCL13", "LAG3", "CD39", ...]}
...
}
Alternatively, SPECTRA can take in a set of adjacency matrices representing networks for each cell type:
gene_set_annotations = {
"global": adjacency_matrix1,
"CD8_T": adjacency_matrix2,
...
}
A pypi package will be available soon. For installation:
git clone https://github.com/dpeerlab/spectra
cd spectra
pip install .
Data used for the examples can be found here:
https://www.dropbox.com/s/jm49egj7lhe83fe/Bassez_10k_subset_scran.h5ad?dl=0
We start by importing spectra and a helper package that estimates the number of factors to use for spectra by bulk eigenvalue matching analysis. In some cases, we can run spectra with number of factors equal to number of gene sets.
from spectra import spectra as spc
from spectra import K_est as kst
L = kst.estimate_L(adata, attribute = "cell_type", highly_variable = True)
model = spc.est_spectra(adata = adata, L = L, gene_set_dictionary = gene_set_dictionary, cell_type_key = "cell_type", use_highly_variable = True)
This latter function stores three important quantities in the AnnData.
factors = adata.uns['SPECTRA_factors'] # factors x genes matrix that tells you how important each gene is to the resulting factors
markers = adata.uns['SPECTRA_markers'] # factors x K [where K is a user specified number] list of K top markers per factor
cell_scores = adata.obsm['SPECTRA_cell_scores'] # cells x factors matrix of cell scores
vocab = adata.var['spectra_vocab'] # boolean matrix of size # of genes that indicates the set of genes used to fit spectra
Model parameters can be accessed with functions associated with the model object
model.return_eta_diag()
model.return_cell_scores()
model.return_factors()
model.return_eta()
model.return_rho()
model.return_kappa()
model.return_gene_scalings()
One outcome of fitting spectra is to fit a gene-gene graph where edges represent similarities between latent variables associated with each gene (a smoothed version of transcriptional similarity) To access this for say, TNK cells; use the following
soft_graph = model.return_graph(ct = "TNK")
for large numbers of genes its clumsy to visualize the whole graph - to visualize a subgraph formed around a particular list of genes, use:
out = spc.graph_network(adata, soft_graph, gene_set)
out.show("test_graph.html")
this will take N closest genes to your gene set and only visualize this subgraph. The interactive graph file gets saved as an html. To visualize multiple gene sets at the same time, we have a different version of the function that assigns a random color to each gene set:
#gene sets is a list of lists
out = spc.graph_network_multiple(adata,soft_graph, gene_sets)
out.show("test_graph.html")
#save trained model
model.save("test_model")
#initialize a model and load trained model
model = SPECTRA_Model(X = X, labels = labels, L = L, vocab = vocab, gs_dict = gene_set_dictionary)
model.load("test_model")
model.__store_parameters()
For smaller problems we can use a memory intensive EM algorithm instead
X = adata.X
A = binary adjacency matrix
model = SPECTRA_EM(X = X, A= A, T = 4)
model.fit()
model.internal_model
SPECTRA(
(theta): ParameterDict(
(global): Parameter containing: [torch.FloatTensor of size 4324x20]
(B): Parameter containing: [torch.FloatTensor of size 4324x31]
(M): Parameter containing: [torch.FloatTensor of size 4324x15]
(Neutrophil): Parameter containing: [torch.FloatTensor of size 4324x3]
(TNK): Parameter containing: [torch.FloatTensor of size 4324x13]
)
(alpha): ParameterDict(
(B): Parameter containing: [torch.FloatTensor of size 6762x51]
(M): Parameter containing: [torch.FloatTensor of size 29328x35]
(Neutrophil): Parameter containing: [torch.FloatTensor of size 3492x23]
(TNK): Parameter containing: [torch.FloatTensor of size 77495x33]
)
(eta): ParameterDict(
(global): Parameter containing: [torch.FloatTensor of size 20x20]
(B): Parameter containing: [torch.FloatTensor of size 31x31]
(M): Parameter containing: [torch.FloatTensor of size 15x15]
(Neutrophil): Parameter containing: [torch.FloatTensor of size 3x3]
(TNK): Parameter containing: [torch.FloatTensor of size 13x13]
)
(gene_scaling): ParameterDict(
(global): Parameter containing: [torch.FloatTensor of size 4324]
(B): Parameter containing: [torch.FloatTensor of size 4324]
(M): Parameter containing: [torch.FloatTensor of size 4324]
(Neutrophil): Parameter containing: [torch.FloatTensor of size 4324]
(TNK): Parameter containing: [torch.FloatTensor of size 4324]
)
)