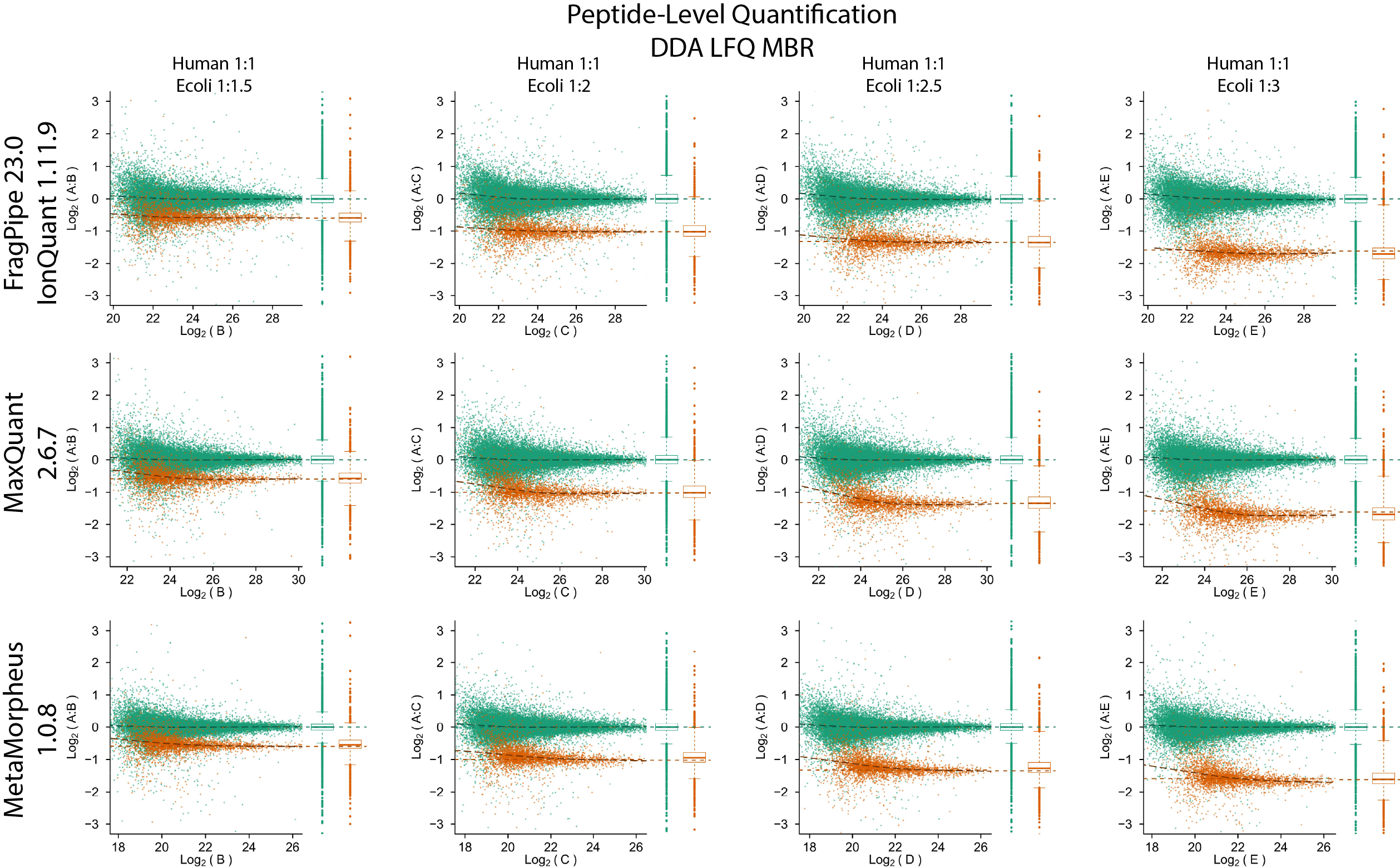
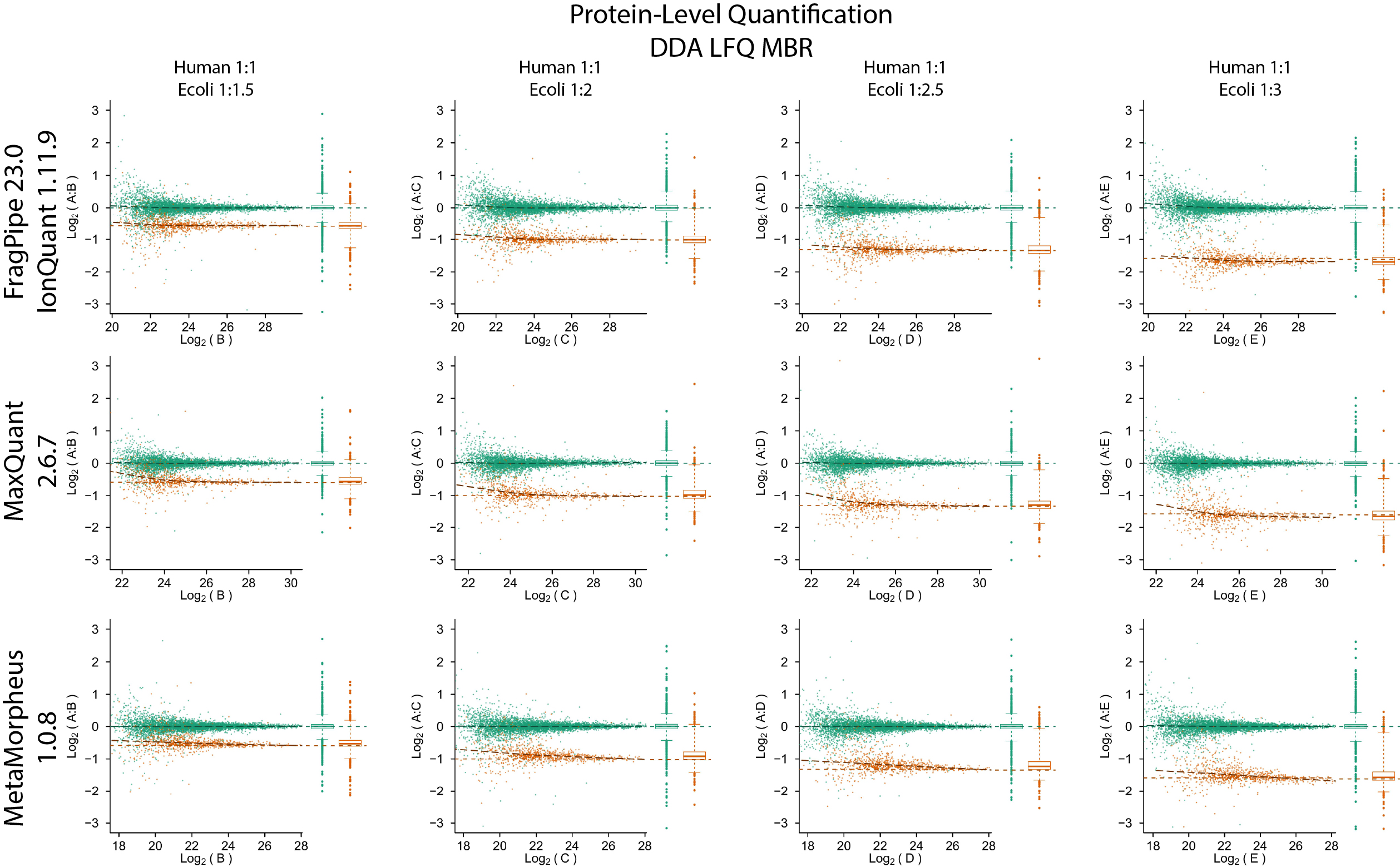
IonQuant is a fast and comprehensive tool for MS1 precursor intensity-based quantification for timsTOF PASEF DDA and non-timsTOF (e.g., Orbitrap) data. It enables label-free quantification with false discovery (FDR) controlled match-between-runs (MBR). It can also be used for quantification in labelling-based experiments such as those involving SILAC, dimethyl, or similar labelling strategies. IonQuant is available as part of FragPipe (recommended option), but can also be run as a command-line tool.
- Java 1.8+.
extfolder from MSFragger.
Note: Bruker's native library needs Visual C++ Redistributable for Visual Studio 2017 in Windows. If you see an error saying cannot find Bruker native library, please try to install the Visual C++ redistibutable.
The latest IonQuant standalone JAR can be downloaded from here.
- Using FragPipe coupled with MSFragger and IonQuant to analyze samples
- Analyzing SILAC (or other labelling coupled with MS1-based quantification) samples with FragPipe
- Running MSstats using MSstats.csv from IonQuant
java -jar IonQuant.jar <options> --specdir <one directory to the spectral files> <.pepXML files>
Options:
--specdir <string> # Directory containing the spectral files (d/mzml/mzxml/raw). One --specdir indicates one spectral directory and can have multiple --specdir.
--threads <integer> # Number of threads. 0 = all logical cores. Default: 0
--mztol <float> # MS1 tolerance in PPM. Default: 10.0
--imtol <float> # 1/K0 tolerance. Default: 0.05
--rttol <float> # Retention time tolerance. Unit: min. Default: 0.4
--seedmz 0/1 # M/Z used as the start point of tracing. 0 = calculated M/Z; 1 = observed M/Z. Default:
--psm <string> # Path to Philosopher's psm.tsv. One --psm indicates one psm.tsv and can have multiple --psm. Optional. Default: <blank>
--multidir <string> # Output directory for the multi-experimental result. Optional. Default: <blank>
--normalization 0/1 # Normalize the intensities across all runs. Default: 1
--minisotopes 1/2/3 # Minimum isotopes required in feature extraction. Default: 2
--minions <integer> # Minimum ions required in quantifying proteins. Default: 2
--excludemods <string> # Excluded modifications in quantifying peptide sequences and proteins. Format: <amino acid><mass>;... Default: <blank>
--proteinquant 1/2 # Protein quantification algorithm. 1 = top-N, 2 = MaxLFQ. Default: 2
--minexps <int> # Minimum experiments in picking an ion for quantifying proteins. Only available when --proteinquant 1. Default: 2
--minfreq <float> # Minimum required frequency of an ion being selected for protein quantification. Only available with --proteinquant 1. Default: 0.5
--tp <int> # Number of ions used in quantifying each protein. If 0, using all ions. Only available with --proteinquant 1. Default: 3
--mbr 0/1 # Perform match-between-runs. Default: 0
--mbrrttol <float> # Retention time tolerance used in match-between-runs. Unit: min. Default: 1.0
--mbrimtol <float> # 1/K0 tolerance used in match-between-runs. Default: 0.05
--mbrtoprun <integer> # Maximum number of donor runs for each acceptor run. Default: 10
--mbrmincorr <float> # Minimum correlation coefficient between a donor run and its acceptor run. Default: 0
--ionmobility 0/1 # The data has ion mobility information or not (for conventional LC-MS data). Default: 0
--ionfdr <float> # Transferred ion false discovery rate threshold. Default: 0.01
--peptidefdr <float> # Transferred peptide false discovery rate threshold. Default: 1
--proteinfdr <float> # Transferred protein false discovery rate threshold. Default: 1
--light <string> # Light labelling mass. Format: <amino acids><mass>;<amino acids><mass>;... Optional. Default: <blank>
--medium <string> # Medium labelling mass. Format: <amino acids><mass>;<amino acids><mass>;... Optional. Default: <blank>
--heavy <string> # Heavy labelling mass. Format: <amino acids><mass>;<amino acids><mass>;... Optional. Default: <blank>
--requantify 0/1 # Re-quantify unidentified feature based on identified feature. Only activate when --light, --medium, or --heavy is not empty. Default: 1
--writeindex 0/1 # Write indexed file on disk for further usage. 0 = no, 1 = yes. Default: 0Note: in some high-performance computing (HPC) servers, you may need to explicitly specify --threads <integer> in case that Java cannot correctly get the logical core number.
Fast quantitative analysis of timsTOF PASEF data with MSFragger and IonQuant
Fengchao Yu, Sarah E. Haynes, Guo Ci Teo, Dmitry M. Avtonomov, Daniel A. Polasky, Alexey I. Nesvizhskii
Molecular & Cellular Proteomics July 2, 2020, mcp.TIR120.002048; DOI: 10.1074/mcp.TIR120.002048