Data and code for ACL 2023 Findings: Aligning Instruction Tasks Unlocks Large Language Models as Zero-Shot Relation Extractors.
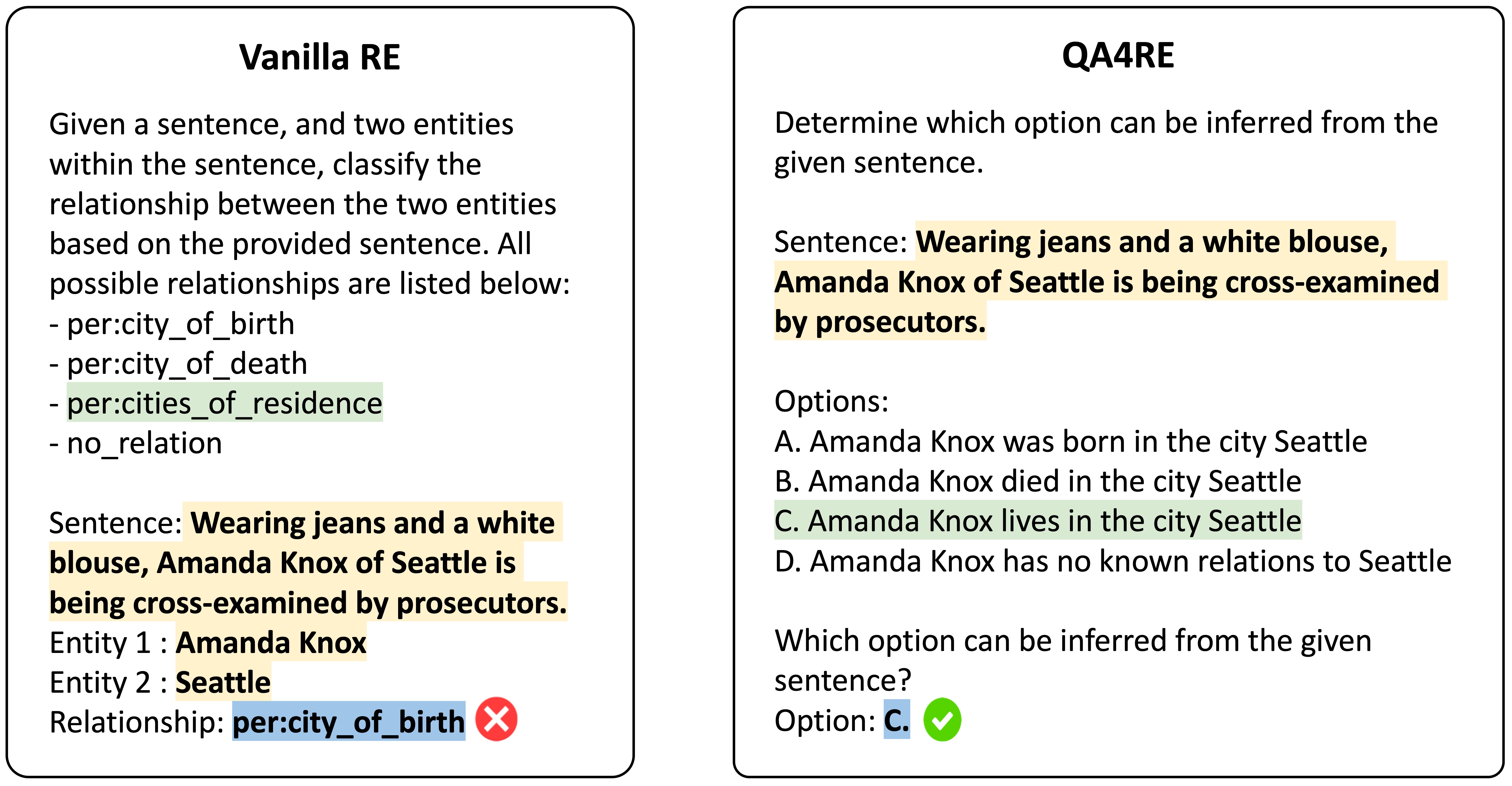
We present LLM-QA4RE, which aligns underrepresented tasks in the instruction-tuning dataset (relation extraction) to a common task (question answering) to unlock instruction-tuned LLMs' abilities on relation extraction.
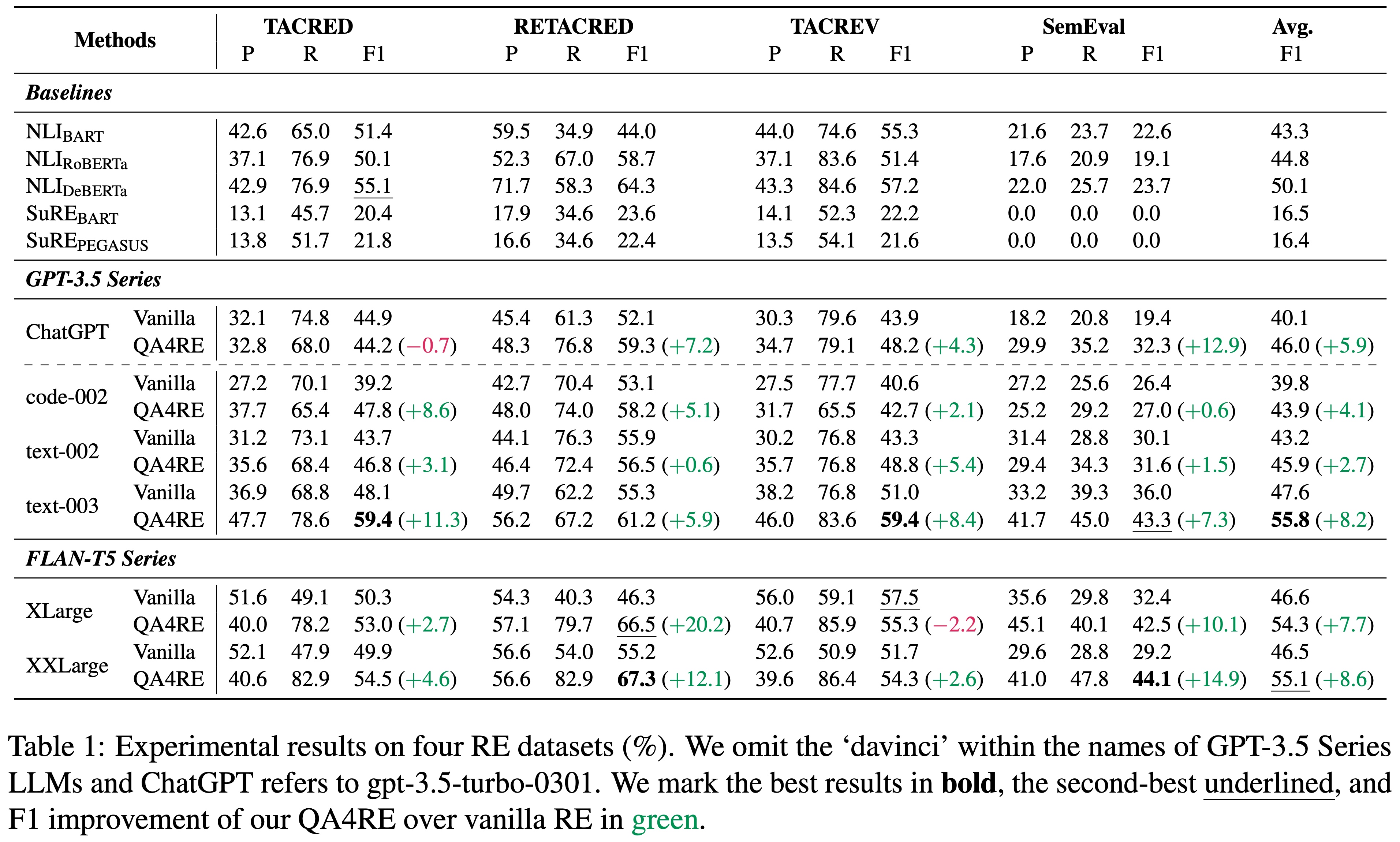
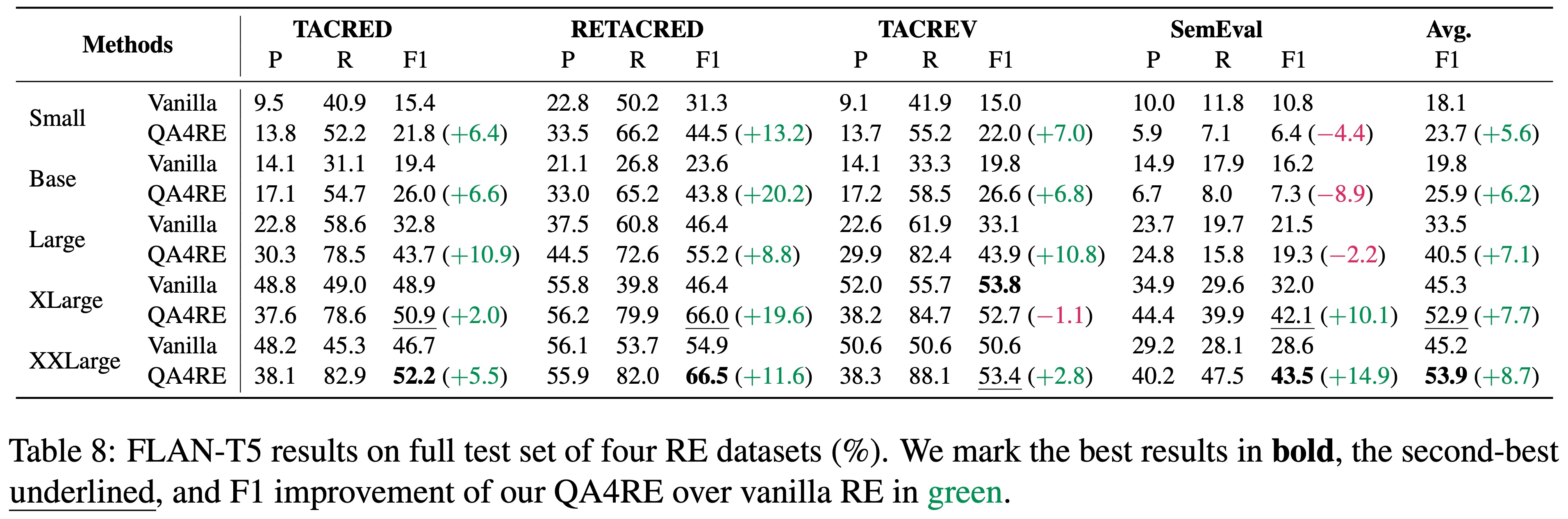
QA4RE achieves significant and consistent performance gains over 6 LLMs across 4 datasets. In addition, it shows strong transferability to model sizes from 175B (GPT-3.5 series) to even 80M (FLAN-T5 Small).
Run the following commands to create a conda environment with the required packages.
conda create -n QA4RE python=3.9.13 pip
conda activate QA4RE
pip install -r requirements.txt
# same env with few-shot-bioIEDownload data and subsets via Google Drive
Results and prompts are saved in Google Drive
Unzip directly in ./ and then the root folder should organize like this:
.
├─── data
│ ├─── RETACRED
│ ├─── TACRED
│ ├─── TACREV
│ ├─── semeval
├─── outputs
│ ├─── RETACRED
│ ├─── TACRED
│ ├─── TACREV
│ ├─── semeval
├─── projs
│ ├─── QA4RE
│ ├─── vanillaRE
│ ├─── README.md
│ ├─── re_templates.py
│ └─── re_utils.py
├─── utils
│ ...
For running, please refer to the README in ./projs dir.
If you find our paper, code, or data helpful, please consider citing the paper:
@inproceedings{Zhang2023LLM-QA4RE,
title={Aligning Instruction Tasks Unlocks Large Language Models as Zero-Shot Relation Extractors},
author={Kai Zhang, Bernal Jiménez Gutiérrez, Yu Su},
booktitle={Findings of ACL 2023},
year={2023}
}
This work is based on our prior work:
@inproceedings{Gutierrez2022Thinking,
title={Thinking about GPT-3 In-Context Learning for Biomedical IE? Think Again},
author={Bernal Jiménez Gutiérrez, Nikolas McNeal, Clay Washington, You Chen, Lang Li, Huan Sun, Yu Su},
booktitle={Findings of EMNLP 2022},
year={2022}
}
If you have any questions, please feel free to contact drogozhang[AT]gmail[DOT]com or open an issue so we can help you better and quicker :)