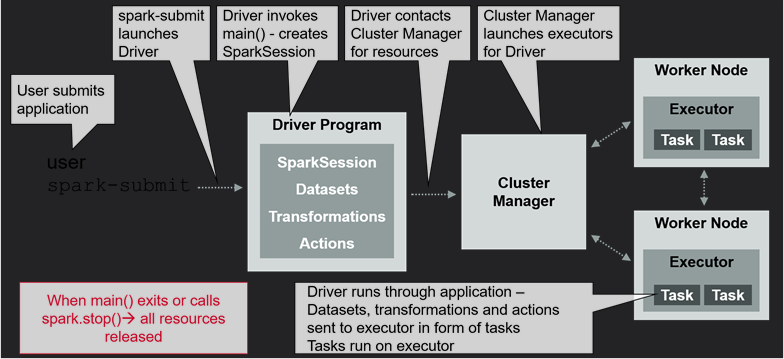
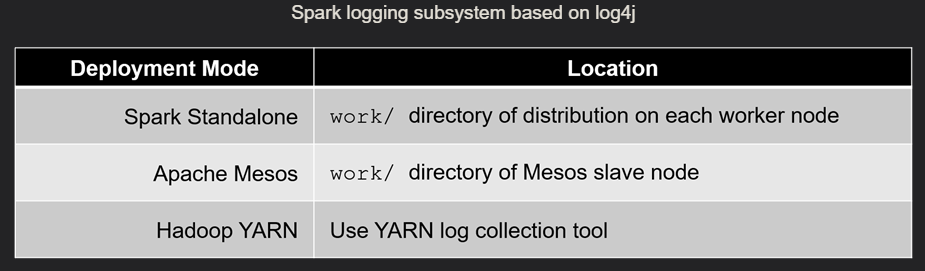
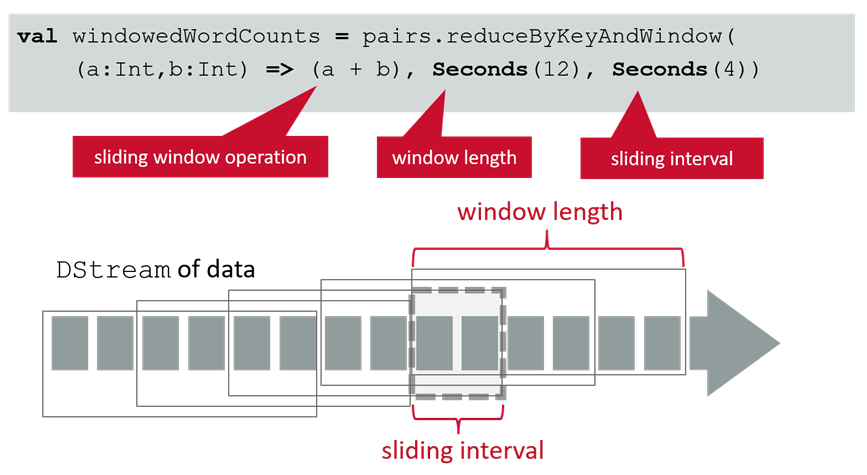
- main documentation
- app examples
- spark by examples
- scala, java, python start point app
- configuration and monitoring
- Mastering apache spark
- spark performance troubleshooting, spark listeners
- spark dynamic resource allocation
- spark trainings
two ways you can improve the performance of your Spark application:
* tuning the degree of parallelism
* avoiding the shuffle of large amounts of data
- local - run in the same VM
- standalone - simple cluster manager
- cluster mode - driver launches on cluster, quit after submission
- client mode - driver launches in the same process, must wait for finish of work
- YARN - reource manager in hadoop
- cluster mode
- client mode
- Mesos - general cluster manager
-
monitoring of execution of SparkJob http://:4040 ( history server )
-
spark properties
new SparkConf().set("spark.executor.memory","1g")bin/spark-submit
--class <main class>
--master <local[n] | spark:<masterurl> | yarn-client/yarn-master | mesos:<mesosurl> >
--deploy-mode <client | cluster>
--conf "spark.driver.extraJavaOptions=-verbose:class"
--conf "spark.executor.extraJavaOptions=-XX:+PrintGCDetails -XX:+PrintGCTimeStamps -verbose:class"
--driver-java-options "-Dlog4j.configuration=file:log4j-local-usage-only.xml" \
--jars <jar1>,<jar2>,<jar3>
< application arguments>
# or
bin/spark-submit
--class <main class>
--master <local[n] | spark:<masterurl> | yarn-client/yarn-master | mesos:<mesosurl> >
--deploy-mode <client | cluster>
--conf "spark.executor.extraJavaOptions=-XX:+PrintGCDetails -XX:+PrintGCTimeStamps"
--driver-java-options "-Dlog4j.configuration=file:log4j-local-usage-only.xml" \
--conf "spark.yarn.appMasterEnv.ENV_VARIABLE_NAME=value_for_master" \
--jars <jar1>,<jar2>
<jar with main>
< application arguments>- conf/spark-env.sh
- log4j.properties
# ability to read files by mask like: /some/path/on/cluster/*/some/*/file
mapreduce.input.fileinputformat.input.dir.recursive=true
spark.rdd.compress=false
# set true for ext4/xfs filesystems
spark.shuffle.consolidateFiles=false
spark.shuffle.spill=true
# memory limit used during reduce by spilling on disk
spark.shuffle.memoryFraction=0.2
spark.shuffle.spill.compress=true
# how much storage will be dedicated to in-memory storage
spark.storage.memoryFraction=0.6
# unrolling serialized data
spark.storage.unrollFraction=0.2
spark.serializer=org.apache.spark.serializer.KryoSerializer
/opt/mapr/spark/spark-2.3.2/bin/spark-submit --verbose --master yarn --deploy-mode cluster ...export SPARK_SUBMIT_OPTS="-Dlog4j.configuration=log4j.properties"
# or
# export SPARK_SUBMIT_OPTS="-Dlog4j.debug=true -Dlog4j.configuration=log4j-local-usage-only.xml"
# or java -D option jvm options
# --driver-java-options "-Xms2G -Doracle.jdbc.Trace=true -Djava.util.logging.config.file=/opt/apache-spark/spark-2.3.0-bin-hadoop2.7/conf/oraclejdbclog.properties -Dcom.sun.management.jmxremote=true -Dcom.sun.management.jmxremote.port=1098 -Dcom.sun.management.jmxremote.authenticate=false -Dcom.sun.management.jmxremote.ssl=false -Djava.net.preferIPv4Stack=true -Djava.rmi.server.hostname=192.168.2.120 -Dcom.sun.management.jmxremote.rmi.port=1095"
/opt/mapr/spark/spark-2.3.2/bin/spark-submit \
--conf "spark.driver.extraJavaOptions=-Dlog4jspark.root.logger=ERROR,console"
--master yarn --deploy-mode cluster \
--queue projects_my_queue \
--name ${USER}_v1-GroundValuesGeneration-${SESSION_ID} \
...spark read log output from yarn
log4j.properties
# Set everything to be logged to the console
log4j.rootCategory=INFO, console
log4j.appender.console=org.apache.log4j.ConsoleAppender
log4j.appender.console.target=System.err
log4j.appender.console.layout=org.apache.log4j.PatternLayout
log4j.appender.console.layout.ConversionPattern=%d{yy/MM/dd HH:mm:ss} %p %c{1}: %m%n
# Settings to quiet third party logs that are too verbose
log4j.logger.org.eclipse.jetty=WARN
log4j.logger.org.eclipse.jetty.util.component.AbstractLifeCycle=ERROR
log4j.logger.org.apache.spark.repl.SparkIMain$exprTyper=INFO
log4j.logger.org.apache.spark.repl.SparkILoop$SparkILoopInterpreter=INFOlog4j.xml
<?xml version="1.0" encoding="UTF-8" ?>
<!DOCTYPE log4j:configuration SYSTEM "log4j.dtd">
<log4j:configuration xmlns:log4j="http://jakarta.apache.org/log4j/">
<appender name="console" class="org.apache.log4j.ConsoleAppender">
<param name="Target" value="System.out"/>
<layout class="org.apache.log4j.PatternLayout">
<param name="ConversionPattern" value="%d{yyyy-MM-dd HH:mm:ss} %-5p %c{1}:%L - %m%n" />
</layout>
</appender>
<logger name="org.apache.spark">
<level value="info" />
</logger>
<logger name="org.spark-project">
<level value="info" />
</logger>
<logger name="org.apache.hadoop">
<level value="info" />
</logger>
<logger name="io.netty">
<level value="info" />
</logger>
<logger name="org.apache.zookeeper">
<level value="info" />
</logger>
<logger name="org">
<level value="info" />
</logger>
<root>
<priority value ="INFO" />
<appender-ref ref="console" />
</root>
</log4j:configuration>list of processes for each executor on separate node
mapr 3810138 152521 0 11:43 ? 00:00:00 /opt/mapr/hadoop/hadoop-2.7.0/bin/container-executor user_a
user_a 3810143 3810138 0 11:43 ? 00:00:00 /bin/bash -c /usr/lib/jvm/java-1.8.0-openjdk-1.8.0.342.b07-1.el7_9.x
user_a 3810284 3810143 2 11:43 ? 00:09:26 /usr/lib/jvm/java-1.8.0-openjdk-1.8.0.342.b07-1.el7_9.x86_64/jre/bin
where 152521 process (parent for container-executor) will be: org.apache.hadoop.yarn.server.nodemanager.NodeManager
import org.apache.spark.sql.SparkSession
val spark = SparkSession.builder.master("local").appName("applicationName").getOrCreate()
spark.read.format("csv").option(....
Coda Hale Metrics Library /conf/metrics.properties
/sbin/start-history-server.sh
- java
- kyro conf.set("spark.serializer", "org.apache.spark.serializer.KryoSerializer") conf.registerKryoClasses(Array(classOf[MyOwnClass1], classOf[MyOwnClass2])) conf.set("spark.scheduler.allocation.file", "/path/to/allocations.xml")
- Ganglia
- OS profiling tools
- JVM utilities
using Alluxio to save/read RDD across SparkContext
java/scala packages
<dependency>
<groupId>org.apache.spark</groupId>
<artifactId>spark-core_2.10</artifactId>
<version>2.2.2</version>
</dependency>
org.apache.spark.SparkContext.*
org.apache.spark.SparkConf
org.apache.spark.api.java.JavaRDD
org.apache.spark.api.java.JavaSparkContext
python
from pyspark import SparkContext, SparkConf
scala
val sparkConfig = new SparkConf().setAppName("my-app").setMaster("local[4]")
val sc = new SparkContext( )
val sqlContext = new org.apache.spark.sql.SQLContext(sc)
val streamingContext = new StreamingContext(conf, Seconds(1))
java
sc = new JavaSparkContext( new SparkConf().setAppName("my-app").setMaster("local[*]") )
new org.apache.spark.sql.api.java.SQLContext(sc)
python
sc = SparkContext( conf = SparkConf().setAppName("my-app").setMaster("local") )
from pyspark.sql import SQLContext
sqlContext = SQLContext(sc)
import spark.implicits._
import org.apache.spark.sql.types._
val customSchema = new StructType(Array( StructField("name", StringType, true), StructType("age", IntegerType, true)))
val dataFrame = spark.read.format("csv").schema(customSchema).load("/path/to/file").toDF("name", "age")
dataFrame.createTempView("viewName")
val dataFrame = spark.read.format("csv").option("inferSchema", true).load("/path/to/file")
.toDF("name", "age")
// name of fields into caseclass must match with name columns in DataFrame
case class Person(name:String, age:Int)
val dataSet = dataFrame.as[Person]
dataSet.map( record=>{ osi3.OsiGroundtruth.GroundTruth.parseFrom(x.osi).getProjString } )
import org.apache.spark.sql.{ DataFrame, SparkSession }
val words:DataFrame = Seq( ("John", 44), ("Mary",38), ("Chak",18)).toDF("name", "age")import spark.implicits._
import org.apache.spark.sql.types._
// create structure
val schema = new StructType()
.add(StructField("name", StringType, true))
.add(StructField("age", IntegerType, true))
// create DataFrame
sqlContext.createDataFrame(
sc.parallelize(
Seq(
Row("John", 27),
Row("Mary", 25)
)
), schema)
// create DataSet
spark.read.format()
val someDF = spark.createDF(
List(
("John", 27),
("Mary", 25)
), List(
("name", StringType, true),
("age", IntegerType, true)
)
)
- load with format
val df=spark.read.format("parquet").load(pathToFile)
// json, parquet, csv
// s3://int-eu-west-1/partitionVersion=v3/vin=xxxxxyyyy/ - read text file
// spark.read.text
// spark.read.textfile
// val spark = SparkSession.builder().getOrCreate()
val df = spark.read.text("/path/to/file/distribution.txt")
df.show()- read jdbc, load jdbc
spark.read.jdbc(url, table, connection_properties)
val jdbcDF = spark.read
.format("jdbc")
.option("url", "jdbc:postgresql:dbserver")
.option("dbtable", "schema.tablename")
.option("user", "username")
.option("password", "password")
.load()
- read csv
import spark.implicits._
val dataFrame = spark.read.csv("/path/to/file")
val dataSet = dataFrame.as[CaseClassName]
dataSet.createTempView("viewName")
- read csv file without header
spark.read.format("csv").option("header", "true").load("/tmp/1.txt")
- read csv file with columns
val df = spark.read.option("inferSchema", true).csv("/path/to/file.csv").toDF("column_1", "column_2", "column_3", "column_4")
df.as[CaseClassMapper]
- read json format
spark.read.format("json").load("/tmp/1.json")
or
context.read.json("my.json")
- read orc format
import org.apache.spark.sql._
val sqlContext = new org.apache.spark.sql.hive.HiveContext(sc)
val data = sqlContext.read.format("orc").load("/path/to/file/*")
or
frame = spark.read.format("orc").load("/path/words.orc")
or
spark.read.orc("/ingestor/46b1-a7d7-c1728088fb48")
- read MapRDB format
import com.mapr.db.spark.sql._
val table1Path="/mapr/zur/vantage/data/store/signal"
val t1 = SparkSessionFunctions(spark).loadFromMapRDB(table1Path)
t1.createOrReplaceTempView("tb1")
// left/inner join
val sqlDF = spark.sql("""
select a.user_id as user_identifier
from tb1 a """)
sqlDF.show()- with format
df.write.format("csv").mode("override").save(filepath)
- csv
df.write.format("csv").save(filepath)
or
df.write.option("header", "true").csv("/data/home/csv")
- json
rdd.write.mode('append').json("/path/to/file")
- orc
data.toDF().write.mode(SaveMode.Overwrite).format("orc").save("/path/to/save/file")
or
dataframe.write.format("orc").mode("overwrite").save("/path/to/file")
- parquet
val usersDF = spark.read.load("users.parquet")
usersDF.select("name", "favorite_color").write.save("namesAndFavColors.parquet")
or
val df=spark.read.format("parquet").load(pathToFile)
df.filter("streamName like 'BN'").show
df.filter("streamName == 'FASETH'").show
df.filter(df("value") === "DAY").show
or
val peopleDF = spark.read.format("json").load("resources/people.json")
peopleDF.select("name", "age").write.format("parquet").save("namesAndAges.parquet")
- jdbc
jdbcDF.write
.format("jdbc")
.option("url", "jdbc:postgresql:dbserver")
.option("dbtable", "schema.tablename")
.option("user", "username")
.option("password", "password")
.save()
- Hive table
dataframe.write.option("path", "/some/path").saveAsTable("some_table_name")
- Solr
// Generate unique key if the 'id' field does not exist
val options = Map("zkhost" -> "134.191.209.235:2181/solr", "collection" -> "label_collection", "gen_uniq_key" -> "true")
dataframe.write.format("solr").options(options).mode(org.apache.spark.sql.SaveMode.Overwrite).save- Elastic
:paste
import spark.implicits._
import org.apache.spark.sql.{ DataFrame, SparkSession }
val words:DataFrame = Seq( ("John", 44), ("Mary",38), ("Chak",18)).toDF("id", "age")
val esHost = new com.bmw.ad.autolabeling.labelstorage.elastic.ElasticSearchHost("https://elasticsearch-stage.stg.addp.com/test2/label");
words.write
.format("org.elasticsearch.spark.sql")
.option("es.nodes.wan.only", "true")
.option("es.net.ssl", "true")
.option("es.net.ssl.truststore.location", "file:///mapr/dp/vantage/deploy/search/trust-store-2022-04-26")
.option("es.net.http.auth.user", "label-s")
.option("es.net.http.auth.pass", "mypassword")
.option("es.port", esHost.getPort)
.option("es.mapping.id", "id")
.option("es.nodes", s"${esHost.getProtocol}://${esHost.getHost}")
.mode("Append")
.save(esHost.getIndexAndType)val dfUnion = df1.union(df2)
val dfUnion = df1.unionAll(df2)words.show()
// words.list()
words.describe()
words.printSchema() val fs = FileSystem.get(sparkSession.sparkContext.hadoopConfiguration)
val path: Path = new Path(pathInHadoopCluster)
if(fs.exists(path)) ...
any task can be executed more than once !!!!
# default value for failing of tasks
spark.task.maxFailures=4
# activate re-launch
spark.speculation = true
# when task is considered like 'slow', when execution time slower than ... median of all executions
spark.speculation.multiplier = 1.5
# default = FIFO
spark.scheduler.mode = FAIR # not to wait in the queue - round robin eviction to be executed somewhere
spark.time(dataFrame.show)
- anonymous functions syntax
(a: Int) => a + 1
- static method
object FunctionCollections{
def sizeCounter(s: String): Int = s.length
}
currentRdd.map(FunctionsCollections.sizeCounter)
- by reference
def myExecutor(rdd: RDD[String]): RDD[String] = {... rdd.map(a=> a+1) }
val data = sc.textFile("some_data.csv")
// debug information of RDD
data.toDebugString()
// descriptions of partitions
dataSet.rdd.partitions.size()
dataSet.rdd.getNumPartitions()
// change number of partitions
dataSet.rdd.repartition(new_size)
dataSet.rdd.coalesce() // optimized version; for decreasing only; doesn't touch part of the data - other parts copy to "untouched"
// statistic values
val statistic = dataFrame.stat
statistic.corr("column1", "column2")
statistic.cov("column1","column2")
statistic.freqItem(Seq("column1"), frequencyValue)
// histogram, mean, stdev, sum, variance, min, max
val myUdf = udf( (s:String)=>(s.trim().substring(s.lastIndexOf('/')) )
// usage example
dataFrame.groupBy(myUdf(dataFrame("column_name"))).show()
usage in SQL
spark.udf.register("trimDirectory", myUdf)
spark.sql("select trimDirectory(column_name) from registeredView groupBy trimDirectory(column_name) ")
any class, that involved into Task should be Serializable
class HelperForString extends Serialization{
}
using lazy initialization for any object inside ( instead of Serializable)
class HelperForString extends Serializable{
@transient lazy val innerHelper = new AnotherHelperForString()
}
Java - default Python - pickle Scala - Kryo
good place for persistence
sc.textFile("data/*.csv").
filter(!_.startsWith("Trip ID")).map(_.split(",")). // skip first line
map(StaticClassUtility.parse(_)).
keyBy(_.column2).
partitionBy(new HashPartitioner(8)).
// after at least one 'heavy' action should be persisted
persist(StorageLevel.xxxxxxx)
rdd1.join(rdd2)
.persist(StorageLevel.xxxxxxx)
* MEMORY_ONLY
* MEMORY_ONLY_SER - compressed version ( CPU consumption )
* MEMORY_AND_DISK
* MEMORY_AND_DISK_SER - compressed version ( CPU consumption )
* DISK_ONLY
cache() == persist() == persist(StorageLevel.MEMORY_ONLY)
// should be called after "action"
rdd.unpersist()
- Bad use case: using SQL DB like key and values ( OLTP style )
select * from table1 where id_key='1234'
select * from table1 where id_key like '%1234%'
- Good use case: using like ExtractTransformLoad ( OLAP style )
- Bad use case: each insert will create new file
insert into table1 select * from table2
- Bad use case: Frequent/Increment updates, using SQL DB for updates
- random access
- delete + insert
- update not working for files
- Good use case: Frequent/Increment updates should be replaced with:
- HDFS + SQL query->Spark
- use 'deleted' column instead of DELETE query ( ClusterBy, Bucketing )
- Bad architecture: HDFS<-Spark<-many users
- Good architecture: HDFS<-Spark<-tableau
- Good for scheduled jobs:
- use dedicated cluster
- output to files
- Bad use case: low latency stream processing:
- send data to queue: JMS, Kafka
val text = sc.textFile("my_own_file")
val counts =
// reduceByKey - reduce all values by key, after - combine the result for each partition
text.flatMap(_.split(" ")).map((_, 1)).reduceByKey(_+_).collectAsMap()
text.flatMap(_.split(" ")).countByValue()
// call reduce( _ + _ ), collect result from each partitions, sent to driver !!! ( only for testing/development )
.countByKey
examples
// approximate result of large dataset:
.countApproxDistinct
// update accumulator with value and return it
.fold((accumulator, value)=> accumulator)
// group all values by key from all partitions into memory !!! ( Memory issue !!!)
.groupByKey
// return all values for specified key
.lookup
// apply map function to value only, avoid shuffling between partitions
.mapValues
- Write ahead logs
write data to fault-tolerant system and send acknoledgement of receipt
- Checkpointing
save data to fault-tolerant system
- Data replication
save data on multiple nodes
- Spark SQL
val employee = sqlContext.sql("select employee.name, employee.age from employee where salary>120000")
employee.map(empl=> "Name:"+empl(0)+" age:"+empl(1).sort(false).take(5).foreach(println)
or
spark.sql("select * from registeredView").show(5)
- Spark Streaming
val lines = streamingContext.socketTextStream("127.0.0.1", 8888)
val words = lines.flatMap(_.split(" "))
words.map(w=>(w,1)) .reduceByKey(_ + _) .print()
- MLib
- GraphX
GraphLoader.edgeListFile(sc, path_to_file)val input = sc.textFile("data/stations/*")
val header = input.first // to skip the header row
val stations = input2.
filter(_ != header2).
map(_.split(",")).
keyBy(_(0).toInt)
stations.join(trips.keyBy(_(4).toInt))-
before start into configuration file "spark-defaults.conf" spark.driver.extraClassPath pathOfJarsWithCommaSeprated
-
during the start ./spark-shell --jars pathOfjarsWithCommaSeprated
-
after start
scala> :require /path/to/file.jar
- multiple lines copy
scala>:paste
...
...
<Ctrl + D># sdk install spark
spark-shell --deploy-mode client --master yarnspark-shell -i /path/to/file.scalacode
object SparkEnterPoint{
def main(args: Array[String]): Unit = {
val spark = SparkSession.builder().appName("traffic-sign-scan").getOrCreate()
import spark.sqlContext.implicits._
println(spark)
...
}
}spark-shell -i <(echo 'val theDate = "my data" ' ; cat <file-name>) spark-shell -i script.scala << END_FILE_MARKER
:quit
END_FILE_MARKERspark-shell \
--jars "/home/some_path/solr-rest_2.11-0.1.jar,/home/someuser/.ivy2/cache/org.json/json/bundles/json-20180813.jar" \
--conf "spark.driver.extraJavaOptions=-agentlib:jdwp=transport=dt_socket,server=y,suspend=y,address=5005"
--conf "spark.executor.extraJavaOptions=-XX:+UseG1GC -XX:+PrintReferenceGC -verbose:gc -XX:+PrintGCDetails -XX:+PrintGCTimeStamps -XX:+PrintAdaptiveSizePolicy -XX:+PrintFlagsFinal -XX:+UnlockDiagnosticVMOptions -XX:+G1SummarizeConcMark"useful changes for local debugging
spark-shell --master local --deploy-mode client \
...- shaded jar
- maven relocations
spark-shell --jars "lightning-1.5.0-SNAPSHOT-3a517e-jar-with-dependencies.jar" \
--name testexecution \
--master yarn \
--deploy-mode client \
--num-executors 100 \
--executor-cores 1 \
--executor-memory 8g \
--driver-memory 8g \
--conf "spark.dynamicAllocation.enabled=false" \
--conf "spark.executor.memoryOverhead=4096" \
--conf "spark.driver.memoryOverhead=4096" \
--conf "spark.executor.extraJavaOptions=-XX:+UseG1GC" \
--conf "spark.driver.extraJavaOptions=-XX:+UseG1GC" \
--conf "spark.serializer=org.apache.spark.serializer.KryoSerializer"
dynamic allocation
-conf "spark.dynamicAllocation.enabled=true" \
-conf "spark.shuffle.service.enabled=true" \
-conf "spark.executor.cores=1" \
-conf "spark.executor.memory=8G" \
-conf "spark.dynamicAllocation.maxExecutors=20" \
-conf "spark.dynamicAllocation.minExecutors=5" \
-conf "spark.dynamicAllocation.initialExecutors=5" \
-conf "spark.dynamicAllocation.executorAllocationRatio=0.5" \
-conf "spark.dynamicAllocation.executorIdleTimeout=60s" \
-conf "spark.dynamicAllocation.schedulerBacklogTimeout=1s" \
-conf "spark.dynamicAllocation.sustainedSchedulerBacklogTimeout=1s" !!! each line in the file should be separate JSON file, minimize each document to one line
spark.read.schema(schema).json(file).select("_corrupt_record").show().
spark.read.json("/home/technik/temp/rdd-processedLabels.json")
sc.textFile("/home/technik/temp/rdd-processedLabels.json").values().map(json.loads)
import scala.util.parsing.json._
sc.wholeTextFiles("/home/technik/temp/rdd-processedLabels.json").values.map(v=>v)
df.head(10).toJSON
df.toJSON.take(10)
// get value from sql.Row
df.map( v=> (v.getAs[Map[String, String]]("processedLabel"), v.getAs[String]("projectName"), v.getAs[String]("sessionId"), v.getAs[Long]("timestamp") ) )
val spark = SparkSession.builder().getOrCreate()
//val sc = spark.sparkContext
//sc.getConf.set("es.port", esHost.getPort).set("es.host", esHost.getHost) DOESN'T TAKE ANY AFFECT
val rdd = spark.read
.format("org.elasticsearch.spark.sql")
.option("es.nodes.wan.only", "true")
.option("es.net.ssl", "true")
.option("es.net.ssl.truststore.location", s"file://$trustStorePath")
.option("es.port", esHost.getPort)
.option("es.mapping.id", "id")
.option("es.nodes", s"${esHost.getProtocol}://${esHost.getHost}")
.load(esHost.getIndexAndType)labelsWithId.write
.format("org.elasticsearch.spark.sql")
.option("es.nodes.wan.only", "true")
.option("es.net.ssl", "true")
.option("es.net.ssl.truststore.location", s"file://$trustStorePath")
.option("es.port", esHost.getPort)
.option("es.mapping.id", "id")
.option("es.nodes", s"${esHost.getProtocol}://${esHost.getHost}")
.mode("Append")
.save(esHost.getIndexAndType):history
:37
:load <path to file>
preparedRdd.take(n).foreach(println)