This repository is part of the CI-SpliceAI software package published in PLOS One.
This is the project comparing different splice prediction tools on variant data. You may also be interested in the code to train CI-SpliceAI, code to use trained models to annotate variants offline, and the website providing online annotation of variants.
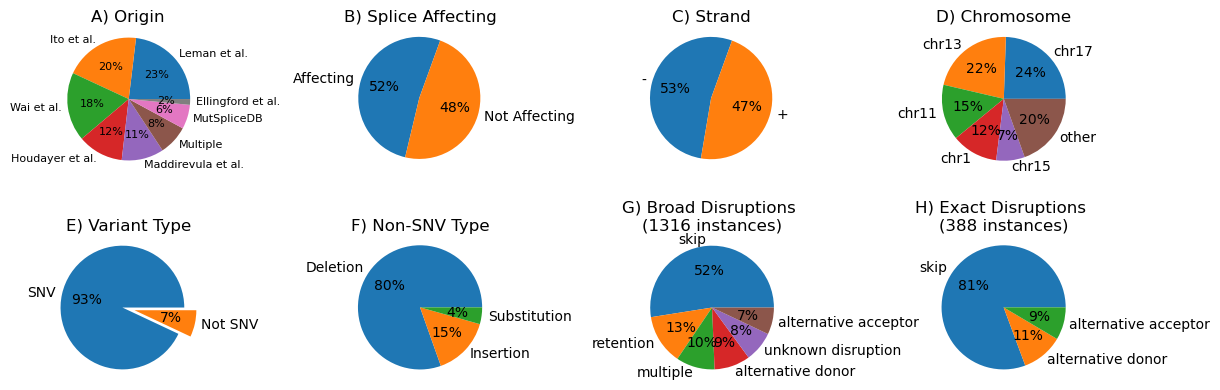
In this project, we are evaluating 6 different splice prediction tools (one of which is ours called CI-SpliceAI) on a corpus of:
- 1,317 variants for a binary affecting/non-affecting task; and
- 388 variants (subset of the first corpus) with annotations of their exact variant effect
This repository contains all variants and all code to re-produce the results obtained.
Visualisations of the variants:
| Algorithm | Coverage | AUC-PR | AUC-ROC | Optimal Threshold | Accuracy |
|---|---|---|---|---|---|
| MES (Sliding) | 100% | 55.68% | 52.97% | 12.5 | 53.42% |
| SQUIRLS | 100% | 91.32% | 91.17% | 0.074 | 85.64% |
| MES (VEP) | 58% | 92.52% | 89.15% | 2.109 | 86.40% |
| MMSplice (Splicing Efficiency) | 99% | 93.03% | 92.56% | 1.119 | 87.23% |
| MMSplice (Pathogenicity) | 99% | 94.13% | 92.84% | 0.961 | 88.53% |
| SpliceAI | 99% | 96.21% | 95.65% | 0.3 | 90.88% |
| CI-SpliceAI | 100% | 97.25% | 96.75% | 0.19 | 92.17% |
| Algorithm | Acceptor Gain | Acceptor Loss | Donor Gain | Donor Loss |
|---|---|---|---|---|
| MES (Sliding) | 0.00% | 1.16% | 2.33% | 2.25% |
| SpliceAI | 87.50% | 77.10% | 79.07% | 78.93% |
| CI-SpliceAI | 93.75% | 78.55% | 79.07% | 82.02% |
These steps were taken:
The variant csv file was parsed into vcf format and normalised (index, normalise rows, align left).
The resulting vcf file is checked in this repository, so you don't need to run the code producing it.
We ran all tools on the vcf file using predict.sh.
Results are checked into predictions/.
Variant data and predictions were analysed and plotted using analysis.sh into analysis/.
This project is built on bash scripts. We suggest running it on a UNIX system; it might be possible to run it on windows using a bash environment like git bash, this is however untested and unsupported.
Before running the setup code, make sure you agree to all licences of third-party components.
Please make sure to install these manual dependencies first:
- Install conda
- Install VEP CLI on Docker and make sure to follow the section linking a volume hosted on ~/vep_data
- Install the MES plugin into your VEP CLI and ensure that MES runs
Then run setup.sh which will automatically:
- Create conda environments with SpliceAI, CI-SpliceAI and MMSplice (through kipoi)
- Download all third party elements like:
- SQUIRLS command line, jannovar annotations, database
- the human reference genome
- GENCODE annotations for MMSplice
- Pre-process GENCODE annotations for MMSplice
This work is licensed under a Creative Commons Attribution 4.0 International License.
By running this code, you are installing third-party software. It is your responsibility to assure that you are following all third party licenses.