Visit http://predfull.com/ to try online prediction
This work was published on Analytical Chemistry: Full-Spectrum Prediction of Peptides Tandem Mass Spectra using Deep Neural Network
Kaiyuan Liu, Sujun Li, Lei Wang, Yuzhen Ye, Haixu Tang
- 2020.05.25: Support predicting non-tryptic peptides
- 2019.09.01: First version
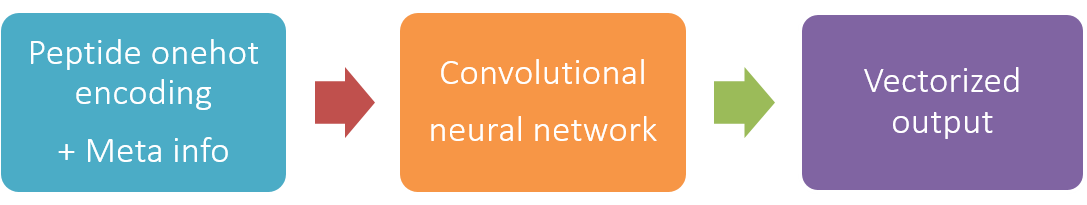
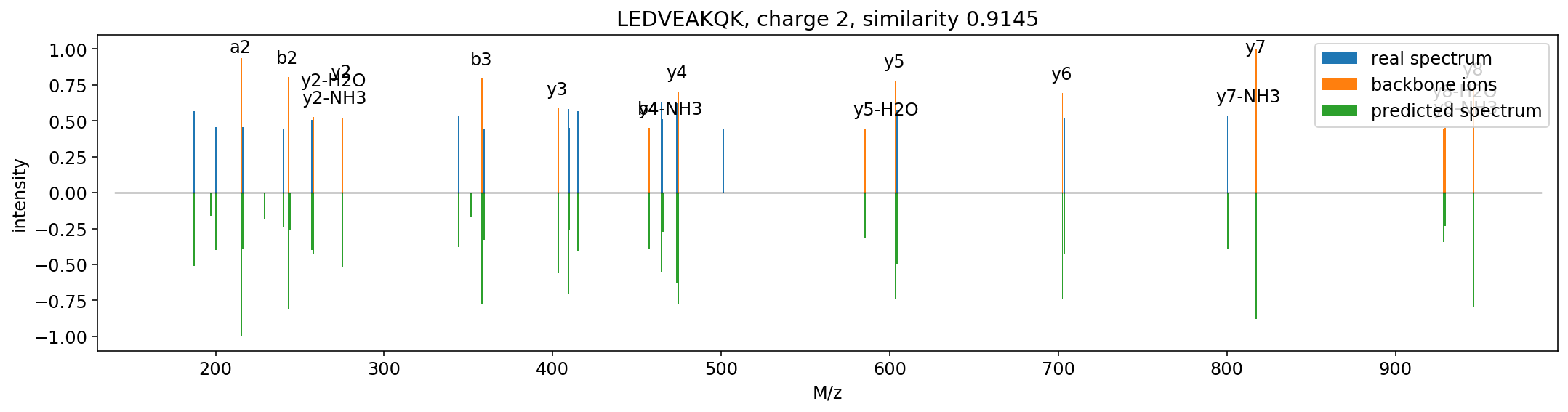
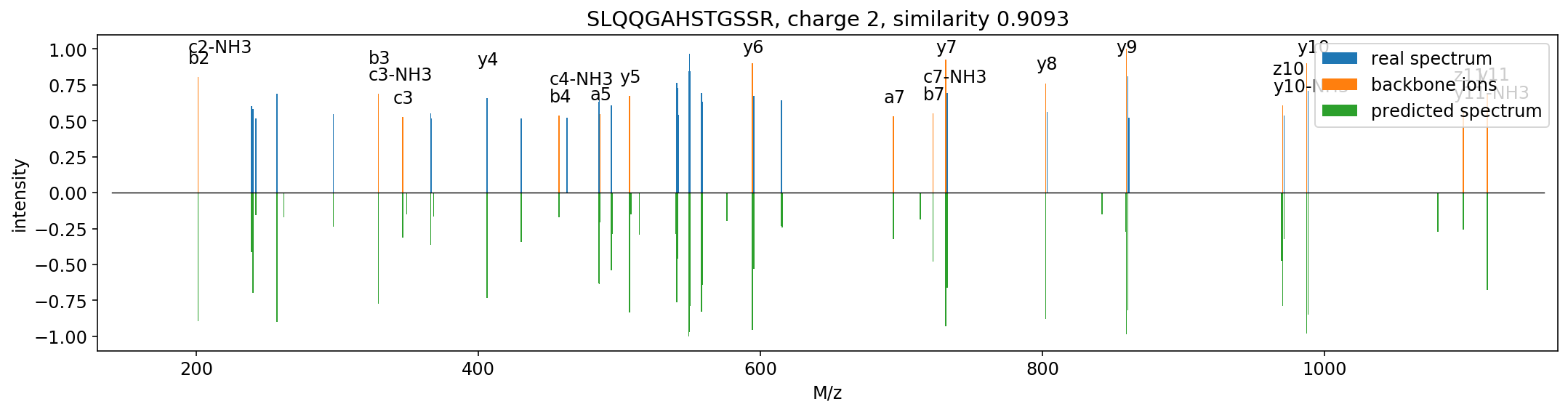
Based on the structure of the residual convolutional networks. Current precision: 0.1 Th.
- This model support only UNMODIFIED peptides (for now, at least)
- This model assume a FIXED carbamidomethyl on C
- The length of input peptides are limited to =< 30
- The prediction will NOT output peaks with M/z > 2000
Recommend to install dependency via Anaconda
- Tensorflow >= 2.0.0
- Pandas >= 0.20
- pyteomics
- lxml
The required input format is TSV, with following columns:
| Peptide | Charge | Type | NCE |
|---|---|---|---|
| AAAAAAAAAVSR | 2 | HCD | 25 |
| AAGAAESEEDFLR | 2 | HCD | 25 |
| AAPAPTASSTININTSTSK | 2 | HCD | 25 |
Apparently, 'Peptide' and 'Charge' columns mean what it says. The 'Type' must be HCD or ETD (in uppercase). NCE means normalized collision energy, set to 25 or 30 if you don't care. Check example.tsv for examples.
Simply run:
python predfull.py --input example.tsv --model pm.h5 --output example.mgf
The output file is in MGF format
- --input : the input file
- --output : the output path
- --model : the pretrained model