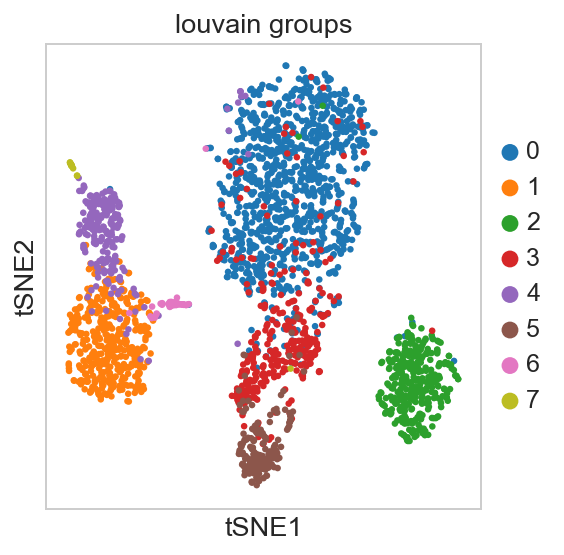
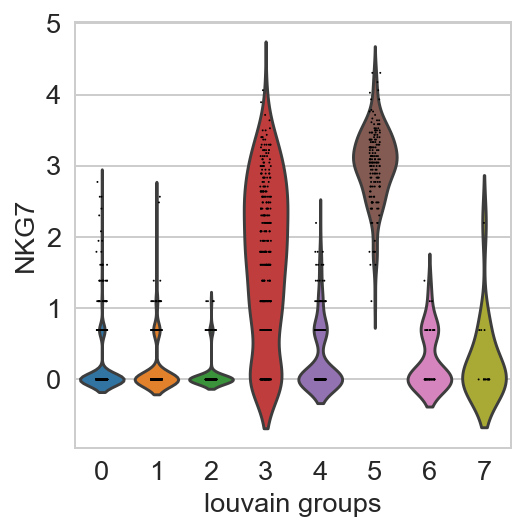
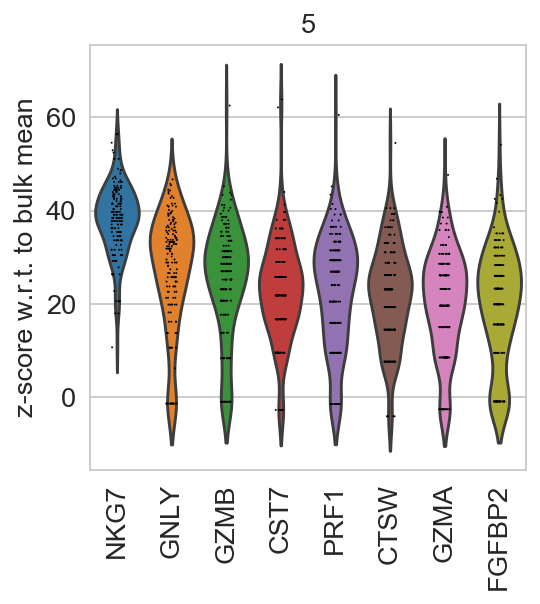
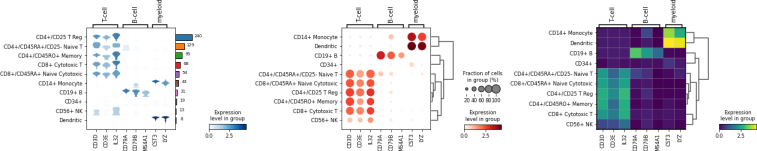
For getting started, we recommend Scanpy’s reimplementation :tutorial:`pbmc3k` of Seurat’s [Satija15]_ clustering tutorial for 3k PBMCs from 10x Genomics, containing preprocessing, clustering and the identification of cell types via known marker genes.
This tutorial shows how to visually explore genes using scanpy. :tutorial:`plotting/core`
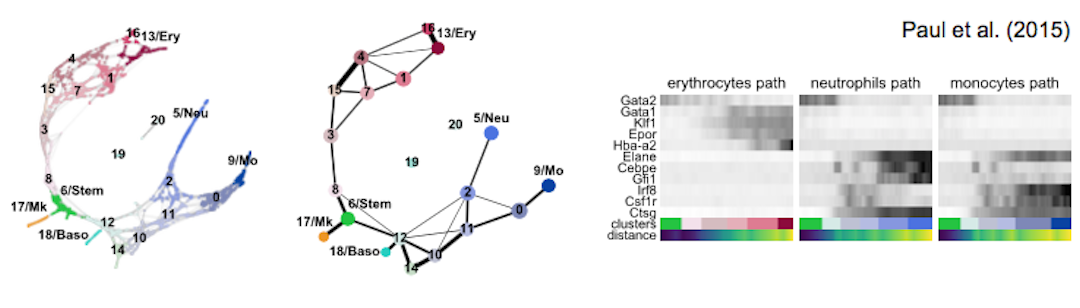
Get started with the following example for hematopoiesis for data of [Paul15]_: :tutorial:`paga-paul15`
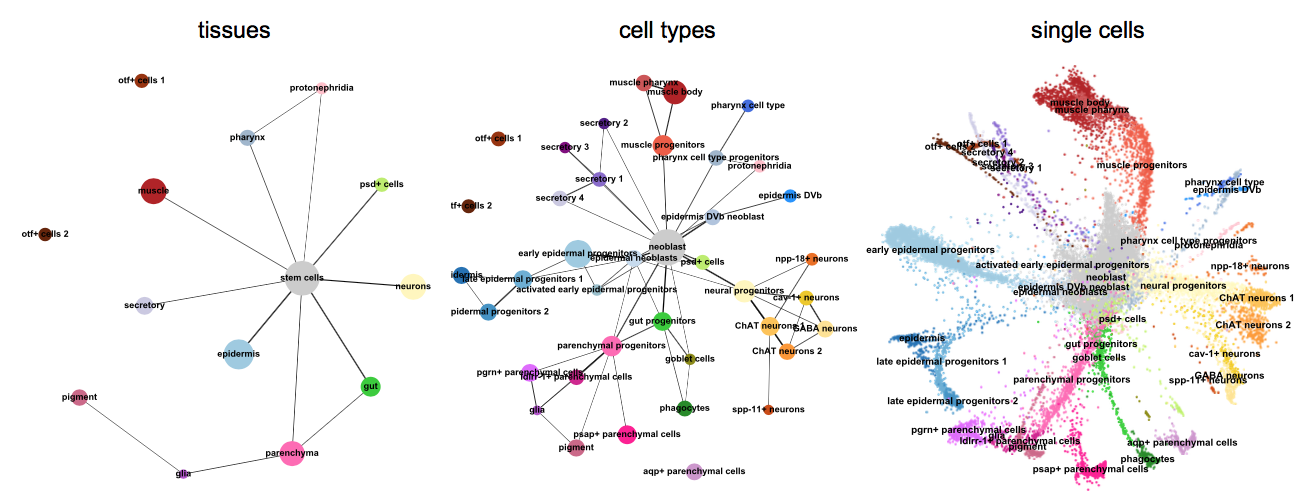
More examples for trajectory inference on complex datasets can be found in the PAGA repository [Wolf19]_, for instance, multi-resolution analyses of whole animals, such as for planaria for data of [Plass18]_.
As a reference for simple pseudotime analyses, we provide the diffusion pseudotime (DPT) analyses of [Haghverdi16]_ for two hematopoiesis datasets: DPT example 1 [Paul15]_ and DPT example 2 [Moignard15]_.
Map labels and embeddings of reference data to new data: :tutorial:`integrating-data-using-ingest`
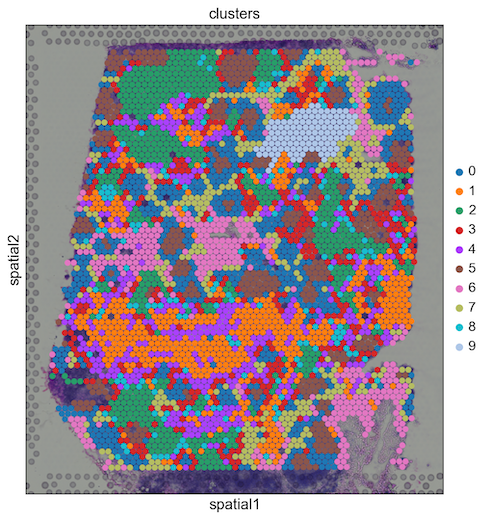
- Basic analysis of spatial data: :tutorial:`spatial/basic-analysis`
- Integrating spatial data with scRNA-seq using scanorama: :tutorial:`spatial/integration-scanorama`
- See Seurat to AnnData for a tutorial on anndata2ri.
- See the Scanpy in R guide for a tutorial on interacting with Scanpy from R.
See the cell cycle notebook.
- Visualize and cluster 1.3M neurons from 10x Genomics.
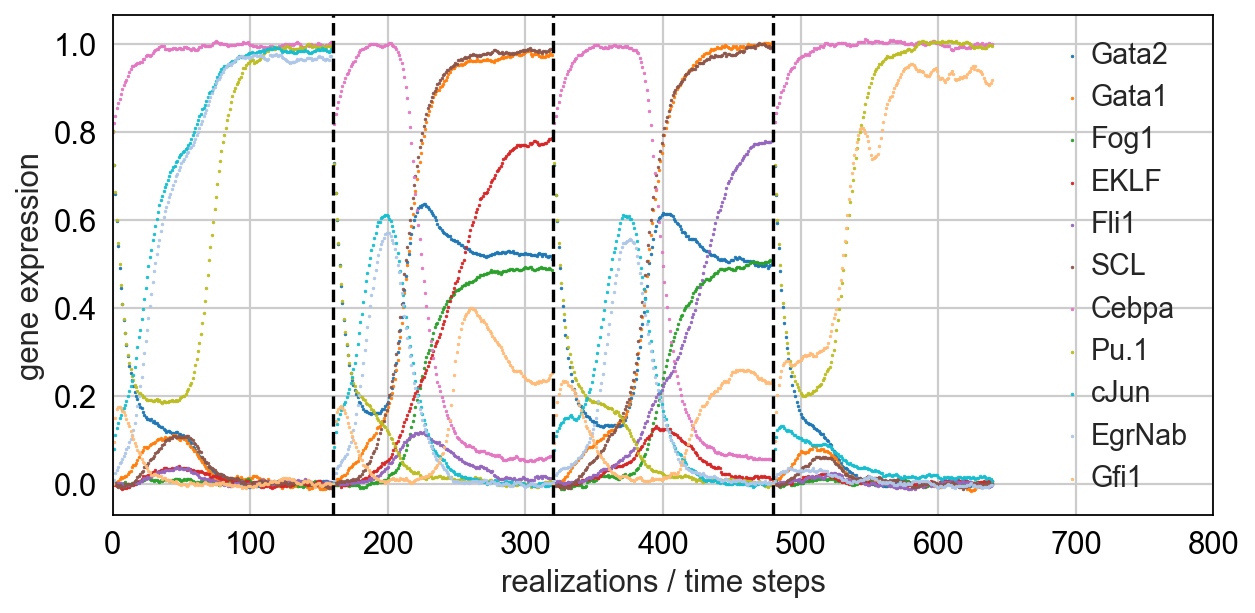
Simulating single cells using literature-curated gene regulatory networks [Wittmann09]_.
- Notebook for myeloid differentiation
- Notebook for simple toggleswitch
See pseudotime-time inference on deep-learning based features for cell cycle reconstruction from image data [Eulenberg17]_.