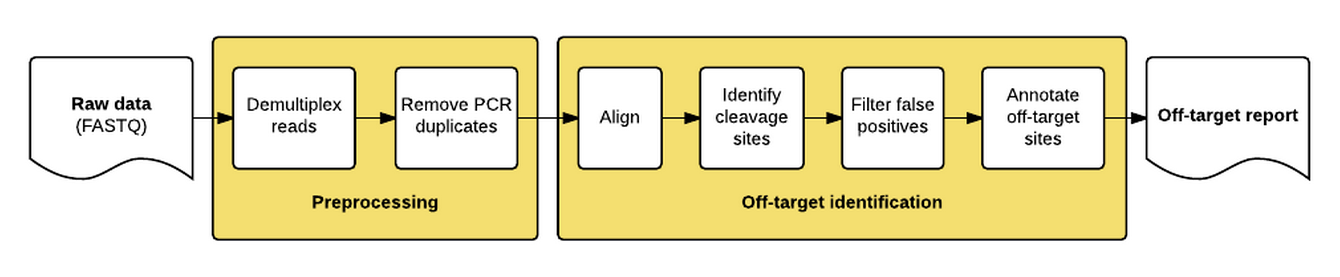
The guideseq package implements our data preprocessing and analysis pipeline for GUIDE-Seq data. It takes raw sequencing reads (FASTQ) as input and produces a table of annotated off-target sites as output.
The package implements a pipeline consisting of a read preprocessing module followed by an off-target identification module. The preprocessing module takes raw reads (FASTQ) from a pooled multi-sample sequencing run as input. Reads are demultiplexed into sample-specific FASTQs and PCR duplicates are removed using unique molecular index (UMI) barcode information.
- Python (2.6, 2.7, or PyPy)
- bwa alignment tool
- bedtools genome arithmetic utility
- Reference genome .fasta file (we recommend hg19)
Using this software is easy, just make sure you have all of the dependencies installed and then grab a copy of this repository.
- Download the
bwaexecutable from their website. Extract the file and make sure you can run it by typing/path/to/bwaand getting the program's usage page. - Download the
bedtoolspackage by following directions from their website. Make sure you can run it by typing/path/to/bedtoolsor justbedtoolsand get the program's usage page. - Make sure you have a copy of a reference genome
.fastafile. We recommend hg19. - Download and extract the
guideseqpackage. You can do this either by downloading the zip and extracting it manually, or by cloning the repositorygit clone --recursive https://github.com/aryeelab/guideseq.git. - Install the
guideseqdependencies by entering theguideseqdirectory and runningpip install -r requirements.txt.
Using this tool is simple, just create a .yaml manifest file referencing the dependencies and sample .fastq.gz file paths. Then, run python /path/to/guideseq.py -m /path/to/manifest.yaml
Below is an example manifest.yaml file:
reference_genome: /Volumes/Media/hg38/hg38.fa
output_folder: ../test/output
bwa: bwa
bedtools: bedtools
undemultiplexed:
forward: ../test/data/undemux.r1.fastq.gz
reverse: ../test/data/undemux.r2.fastq.gz
index1: ../test/data/undemux.i1.fastq.gz
index2: ../test/data/undemux.i2.fastq.gz
samples:
control:
target:
barcode1: CTCTCTAC
barcode2: CTCTCTAT
description: Control
EMX1:
target: GAGTCCGAGCAGAAGAAGAANGG
barcode1: TAGGCATG
barcode2: TAGATCGC
description: Round 3 Adli
Absolute paths are recommended. Be sure to point the bwa and bedtools paths directly to their respective executables.
Once you have a manifest file created, you can simply execute python PATH/TO/guideseq.py -m PATH/TO/MANIFEST.YAML to run the entire pipeline. All output files, including the results of each individual step, will be placed in the output_folder.
[License Information]
[Disclaimer]