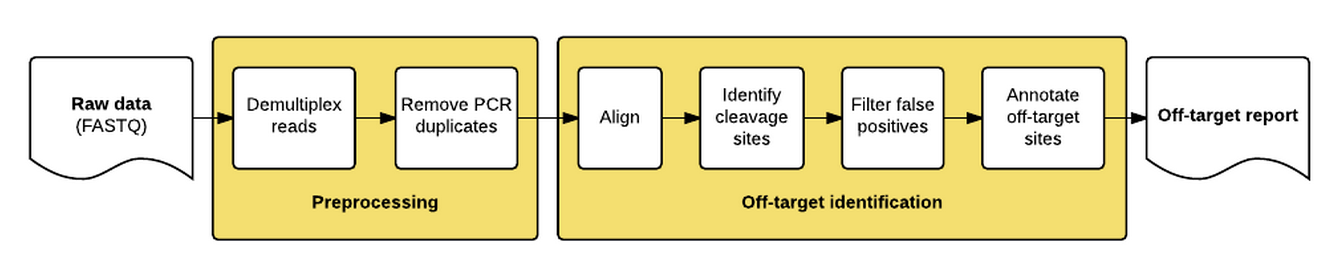
The guideseq package implements our data preprocessing and analysis pipeline for GUIDE-Seq data. It takes raw sequencing reads (FASTQ) as input and produces a table of annotated off-target sites as output.
The package implements a pipeline consisting of a read preprocessing module followed by an off-target identification module. The preprocessing module takes raw reads (FASTQ) from a pooled multi-sample sequencing run as input. Reads are demultiplexed into sample-specific FASTQs and PCR duplicates are removed using unique molecular index (UMI) barcode information.
This package also produces visualizations of detected off-target sites, as seen below.
- Python (2.6, 2.7, or PyPy)
- bwa alignment tool
- bedtools genome arithmetic utility
- Reference genome fasta file (Example)
Using this software is easy, just make sure you have all of the dependencies installed and then grab a copy of this repository.
- Download the
bwaexecutable from their website. Extract the file and make sure you can run it by typing/path/to/bwaand getting the program's usage page. - Download the
bedtoolspackage by following directions from their website. Make sure you can run it by typing/path/to/bedtoolsor justbedtoolsand get the program's usage page. - Make sure you have a copy of a reference genome fasta file. (Example)
- Download and extract the
guideseqpackage. You can do this either by downloading the zip and extracting it manually, or by cloning the repositorygit clone --recursive https://github.com/aryeelab/guideseq.git. - Install the
guideseqdependencies by entering theguideseqdirectory and runningpip install -r requirements.txt.
Using this tool is simple, just create a .yaml manifest file referencing the dependencies and sample .fastq.gz file paths. Then, run python /path/to/guideseq.py all -m /path/to/manifest.yaml
Below is an example manifest.yaml file:
reference_genome: /Volumes/Media/hg38/hg38.fa
output_folder: ../test/output
bwa: bwa
bedtools: bedtools
undemultiplexed:
forward: ../test/data/undemux.r1.fastq.gz
reverse: ../test/data/undemux.r2.fastq.gz
index1: ../test/data/undemux.i1.fastq.gz
index2: ../test/data/undemux.i2.fastq.gz
samples:
control:
target:
barcode1: CTCTCTAC
barcode2: CTCTCTAT
description: Control
EMX1:
target: GAGTCCGAGCAGAAGAAGAANGG
barcode1: TAGGCATG
barcode2: TAGATCGC
description: Round 3 Adli
Absolute paths are recommended. Be sure to point the bwa and bedtools paths directly to their respective executables.
Once you have a manifest file created, you can simply execute python PATH/TO/guideseq.py all -m PATH/TO/MANIFEST.YAML to run the entire pipeline. All output files, including the results of each individual step, will be placed in the output_folder.
You can also run each step of the pipeline individually by running python PATH/TO/guideseq.py [STEP] [OPTIONS]. Supported commands are:
all: Run all pipeline steps (manifest required)demultiplex: Demultiplex undemultiplexed files (manifest required)umitag: UMI-tag demultiplexed filesconsolidate: Consolidate UMI-tagged filesalign: Align consolidated reads to a reference genomeidentify: Identify offtarget sites from aligned readsfilter: Filter identified background sites from identified treatment sitesvisualize: Produce visualization of off-target sites from result of theidentifystep
To run tests, you must first create a .genome text file in the guideseq root folder with a single line containing the absolute path to the hg38 reference genome .fasta file. Then, you can simply run tox to run the full test pipeline.
This software is licensed under the GNU AGPL license. For usage information about this license, see the GNU AGPL information page.