We provided command-line versions that can be used to design the cytosine base editor mediated gene inactivation for large amounts of CDS.
We developed the 'autocbei' to automate the calculation of potential CBEI loci of the target CDSs and to perform statistics and calculations.
"autocbei" relies on python3 and requires 'biopython' and 'matplotlib' packages. The current version (1.6.3) supports ‘conda’ and ‘pip’ installations.
If Python3 (Python >= 3.6.0) is already included in your operating system, autocbei can be automatically installed via pip.
pip install autocbei
#or Upgrade
pip install --upgrade autocbeiUse the following command to determine which version of Python you have installed:
python -V
If you want to try out the latest but unreleased version (possibly unstable) of autocbei, please follow these commands:
# Download CRISPR-CBEI from Github
1. git clone https://github.com/atlasbioinfo/CRISPR-CBEI.git
# Install biopython and matplotlib
2. conda install -y biopython matplotlib
# Run autocbei and show help info
3. python run_autocbei.py -hIf the installation of "autocbei" has been successful, the sample file is located at [PYTHON_HOME]/site-packages/autocbei/Bacillus_subtilis.part500.cds.all.fa
If installed with Conda, it should be located at:
~/anaconda3/envs/autocbei/lib/python3.7/site-packages/autocbei/Bacillus_subtilis.part500.cds.all.fa
Or, it can be downloaded via "https://github.com/atlasbioinfo/CRISPR-CBEI/blob/master/autocbei/Bacillus_subtilis.part500.cds.all.fa".
The -h parameter shows help information.
autocbei -h
usage: autocbei [-h] [-ns] [-o OUTPREFIX] CDS.fasta
Enter fasta file of CDSs, output base editor's potential editing site and
statistics information. The demo CDS Fasta file: "[PATHON_HOME]/site-
packages/autocbei/Bacillus_subtilis.part500.cds.all.fa".
positional arguments:
CDS.fasta CDSs in fasta format.
optional arguments:
-h, --help show this help message and exit
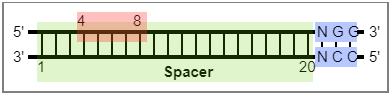
-bef BEF [Optional] The file containing the base editor definition, seperate with ",". eg: "BE,NGG,20,4,8,5", that
is, "name, PAM, spacer length, edit beg, edit end, PAM at 5' or 3' of spacer". 13 base editors are included
by default.
-ns, --nostat Only run CBEI design without statistics and plot.
-o OUTPREFIX, --outprefix OUTPREFIX
Directory prefixes can be customized. Default: "CBEI"
(CBEIRaw, CBEIPlot, CBEIRes).
Calculate the potential CRISPR base editing site information of CDSs
autocbei Bacillus_subtilis.part500.cds.all.fa
Calculation only without statistics and ploting:
autocbei -ns Bacillus_subtilis.part500.cds.all.fa -o Bac
Customize base editors
autocbei -bef exampleBE.csv Bacillus_subtilis.part500.cds.all.fa
cat exampleBE.csv
#sep with comma ","
#BE,NGG,20,4,8,5
#YE1-BE3,NGG,20,5,7,5
#name, PAM, spacer length, edit beg, edit end, PAM at 5' or 3' of spacer
The base editors example files can be found here: https://github.com/atlasbioinfo/CRISPR-CBEI/blob/master/autocbei/exampleBE.csv
The autoCBEI contains 13 commonly used base editors.
#Set the Base editor parameter
# [PAM, spacer length, edit beg, edit end, direction]
# Direction refers to spacer at the 5' or 3' end of PAM sequence (5 or 3, respectively).
beinfos = {
"BE":["NGG",20,4,8,5],
"YE1-BE3":["NGG",20,5,7,5],
"EE-BE3":["NGG",20,5,6,5],
"YEE-BE3":["NGG",20,6,6,5],
"VQR-BE3":["NGAN",20,4,11,5],
"VRER-BE3":["NGCG",20,3,10,5],
"SaBE":["NNGRRT",21,3,12,5],
"Sa(KKH)-BE3":["NNNRRT",21,3,12,5],
"Cas12a–BE":["TTTV",20,10,12,3],
"Target-AID":["NGG",20,2,4,5],
"Target-AID-NG":["NG",20,2,4,5],
"xBE3":["NG",20,4,8,5],
"BE-PLUS":["NGG",20,4,14,5]
# Or, add your own Base editors
# "BE name" :[PAM, SpLength, EditBeg, EditEnd, Direction]
}PAM: PAM sequence.
SpLength: Spacer length.
EditBeg: Edit windows begin.
EditEnd: Edit windows end.
Direction: 5 or 3. Spacer is at the 5 'end or 3' end of the PAM sequence. The example in the figure is the 5.
The output message of simply run
autocbei Bacillus_subtilis.part500.cds.all.fashould be:
Input file: Bacillus_subtilis.part500.cds.all.fa
Output directory: CBEIRaw
Base editors:
BE PAM Spacer EditBegin EditEnd Direction
BE NGG 20 4 8 5
YE1-BE3 NGG 20 5 7 5
EE-BE3 NGG 20 5 6 5
YEE-BE3 NGG 20 6 6 5
VQR-BE3 NGAN 20 4 11 5
VRER-BE3 NGCG 20 3 10 5
SaBE NNGRRT 21 3 12 5
Sa(KKH)-BE3 NNNRRT 21 3 12 5
Cas12a–BE TTTV 20 10 12 3
Target-AID NGG 20 2 4 5
Target-AID-NG NG 20 2 4 5
xBE3 NG 20 4 8 5
BE-PLUS NGG 20 4 14 5
Start calculating: BE
CBEI calculation of base editor "BE" , done!
Start calculating: YE1-BE3
CBEI calculation of base editor "YE1-BE3" , done!
Start calculating: EE-BE3
CBEI calculation of base editor "EE-BE3" , done!
Start calculating: YEE-BE3
CBEI calculation of base editor "YEE-BE3" , done!
Start calculating: VQR-BE3
CBEI calculation of base editor "VQR-BE3" , done!
Start calculating: VRER-BE3
CBEI calculation of base editor "VRER-BE3" , done!
Start calculating: SaBE
CBEI calculation of base editor "SaBE" , done!
Start calculating: Sa(KKH)-BE3
CBEI calculation of base editor "Sa(KKH)-BE3" , done!
Start calculating: Cas12a–BE
CBEI calculation of base editor "Cas12a–BE" , done!
Start calculating: Target-AID
CBEI calculation of base editor "Target-AID" , done!
Start calculating: Target-AID-NG
CBEI calculation of base editor "Target-AID-NG" , done!
Start calculating: xBE3
CBEI calculation of base editor "xBE3" , done!
Start calculating: BE-PLUS
CBEI calculation of base editor "BE-PLUS" , done!
Calculate complete!
Begin statistics...
The statistics directory: CBEIStat
The plot directory: CBEIPlot
####################
Pie charts for different BEs have been generated.
Path:
CBEIPlot/[BE names]_Bacillus_subtilis.statPie.png
Transcript statistics and mapping completed.
Path:
CBEIPlot/Bacillus_subtilis_CDSlength.png
CBEIPlot/Bacillus_subtilis_GCstats.png
CBEIPlot/Bacillus_subtilis_CodonUsage.png
CBEIPlot/Bacillus_subtilis_CDSlength.png
The comparison of CBEI ratio of different BE has been completed.
Path:
CBEIPlot/Bacillus_subtilis.statBar.png
CBEIPlot/Bacillus_subtilis.statROC.png
CBEI statistics complete
The output directory or files shold be:
├── CBEIPlot
│ ├── BE-PLUS_Bacillus_subtilis.statPie.png
│ ├── BE_Bacillus_subtilis.statPie.png
│ ├── Bacillus_subtilis.statBar.png
│ ├── Bacillus_subtilis.statROC.png
│ ├── Bacillus_subtilis_CDSlength.png
│ ├── Bacillus_subtilis_CodonUsage.png
│ ├── Bacillus_subtilis_GCstats.png
│ ├── ...
├── CBEIRaw
│ ├── BE-PLUS_Bacillus_subtilis.part500.cds.all.fa.cbei
│ ├── BE_Bacillus_subtilis.part500.cds.all.fa.cbei
│ ├── ...
├── CBEIStat
│ ├── BE-PLUS_Bacillus_subtilis.Thre025.tsv
│ ├── BE-PLUS_Bacillus_subtilis.Thre05.tsv
│ ├── BE-PLUS_Bacillus_subtilis.Thre075.tsv
│ ├── ...
KDE22635 Minus 0.8674698795180723 {TGC,[TTT,C(C->T)]A,TAA,GAT,TAA,AA}|T,G TGCTTTCCATAAGATTAAAA 222-203 216,215 219-215 TG 201-202 TC,CC
- Fasta title: E.g., lacZ
- Strand: E.g., Plus
- Relative position: Edit position/Gene length. E.g., 0.012032520325203253
- Rich info: E.g., GT{C,GT[T,TTA,(C->T)]AA,CGT,CGT,GAC,T}|GG,G
- Spacer: E.g., CGTTTTACAACGTCGTGACT
- Spacer position: E.g., 30-49
- Edit position: E.g., 37
- Edit windows: E.g., 33-37
- PAM:E.g., GGG
- PAM position: E.g., 50-52
- Edit pattern: The edit pattern indicated the adjacent nucleotide at 5’ of the editable cytosine. Typically, the in vitro activity of the base editors follows TC ≥ CC ≥ AC > GC. E.g., AC
Filter according to different thresholds.
- Thre25: CBEI sites at the upsteam 25% of the CDS.
- Thre50: CBEI sites at the upsteam 50% of the CDS.
- Thre75: CBEI sites at the upsteam 75% of the CDS.
#Editable sites in the first 50% of the transcript
#Transcript Strand Position CBEIdetail Spacer SpacerRegion EditPosition EditWindowsRegion PAM PAMregion Pattern
KDE22636 Plus 0.3686274509803922 GG{A,TT[T,CTT,(C->T)]AA,CAA,ACA,GTA,C}|TG,G ATTTCTTCAACAAACAGTAC 87-106 94 90-94 TGG 107-109 TC
Pie charts for different BEs have been generated.
Path:
./CBEIPlot/[BE names]_Bacillus_subtilis.statPie.tiff
Transcript statistics and mapping completed.
Path:
./CBEIPlot/Bacillus_subtilis_CDSlength.tiff
./CBEIPlot/Bacillus_subtilis_GCstats.tiff
./CBEIPlot/Bacillus_subtilis_CodonUsage.tiff
./CBEIPlot/Bacillus_subtilis_CDSlength.tiff
The comparison of CBEI ratio of different BE has been completed.
Path:
./CBEIPlot/Bacillus_subtilis.statBar.tiff
./CBEIPlot/Bacillus_subtilis.statROC.tiff
CBEI statistics complete