| title | title_short | tags | authors | affiliations | date | cito-bibliography | event | biohackathon_name | biohackathon_url | biohackathon_location | group | git_url | authors_short | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
An evaluation of EDAM coverage in the Tools Ecosystem and prototype integration of Galaxy and WorkflowHub systems |
An evaluation of EDAM coverage in the Tools Ecosystem and prototype integration of Galaxy and WorkflowHub systems |
|
|
|
13 February 2023 |
paper.bib |
BH22EU |
BioHackathon Europe 2022 |
Paris, France, 2022 |
Project 25 |
Lucie Lamothe, Jennifer Rugaard Bregndahl Jensen \emph{et al.} |
The Tools Ecosystem is a centralized repository for the open and transparent exchange of metadata about software tools and services in bioinformatics and life sciences. It serves as a foundation for the sustainability and interoperability of the diverse Tools Platform services: bio.tools [@usesDataFrom:Ison2019], BioContainers [@usesDataFrom:10.1093/bioinformatics/btx192], OpenEBench [@usesDataFrom:openebench], Bioconda [@usesDataFrom:bioconda], WorkflowHub [@usesDataFrom:workflowhub], usegalaxy.eu [@usesDataFrom:galaxy]. It also includes a number of related resources outside of the ELIXIR Tools Platform (e.g. Debian Med, biii.eu).
Here we report the results of a project started at the BioHackathon Europe 2022. Its goals were to:
- Cross-compare and analyze the metadata centralized in the Tools Ecosystem, including coverage of the EDAM ontology [@usesDataFrom:10.1093/bioinformatics/btt113], and
- Explore methods for connecting tools used in registered Galaxy workflows (i.e. WorkflowHub entries) to the annotations available in bio.tools.
This report is separated into three sections. In the first, we present the results of the two analyses described above. The second section details the methods we used, and finally we discuss potential perspectives for both improved monitoring and curation of the Tools Ecosystem metadata and EDAM, as well as for improving the connectivity and integration between elements of the Ecosystem (i.e. bio.tools, WorkflowHub) and platform services that make use of this Ecosystem (e.g. Galaxy [@usesDataFrom:10.1093/bioinformatics/btt199]).
Here we assess the completeness of the annotation of bio.tools entries with EDAM concepts. The two figures 1a and 1b represent the respective proportions of entries annotated with EDAM topics, operations, data and formats.
 {width=90%}
Figure 1a: bio.tools entries annotation represented as a Venn diagram. Each set here represents the proportion of entries annotated with EDAM topics, operations and data. Overlap areas indicate the proportion of bio.tools entries annotated with e.g. both topics and operations, or topics, operations, and data.
{width=90%}
Figure 1a: bio.tools entries annotation represented as a Venn diagram. Each set here represents the proportion of entries annotated with EDAM topics, operations and data. Overlap areas indicate the proportion of bio.tools entries annotated with e.g. both topics and operations, or topics, operations, and data.
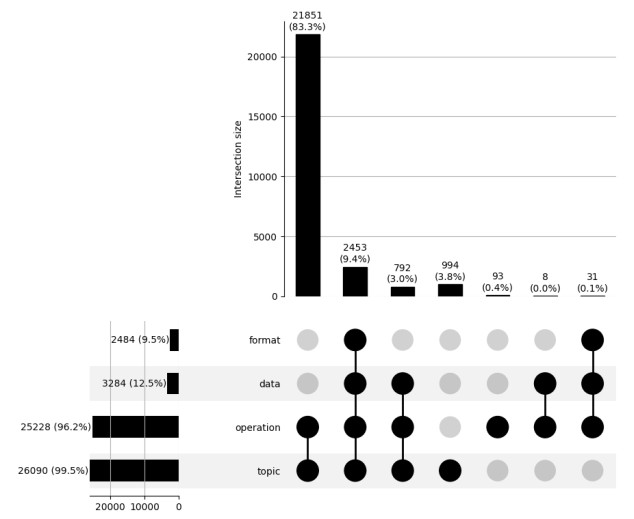 {width=100%}
Figure 1b: bio.tools entries annotation represented as an UpSet plot. Each line represents the proportion of entries annotated with EDAM topics, operations, data and formats. Columns indicate the proportion of bio.tools entries for each combination of intersections (e.g. proportion of entries annotated with topics and operations).
{width=100%}
Figure 1b: bio.tools entries annotation represented as an UpSet plot. Each line represents the proportion of entries annotated with EDAM topics, operations, data and formats. Columns indicate the proportion of bio.tools entries for each combination of intersections (e.g. proportion of entries annotated with topics and operations).
These results show that most entries (83.3%) are annotated with topics and operations, and another significant portion (9.4%) also includes data and format concepts. Only a very small proportion of the entries (1.1%) do not include any EDAM annotation. The explanation for the important proportion of annotated entries is probably that EDAM topics and operations can be specified either by human contributors and curators, but also as the result of an automated text-mining process [@describes:jaaniso2016automatic]. This level of annotation, with 96.2% of the tools including operation annotations, means that for almost all entries the scientific functions of the tools is described.
The analyses in this section assess the usage of EDAM concepts in bio.tools for each of its four sections. We first evaluate the proportion of the EDAM concepts used to annotate entries, and then relate their usage to their suitability for annotation: some EDAM concepts, obsolete or auxiliary, should not be used for annotations.
This usage analysis has been performed with two versions of EDAM: the last stable release (1.25) currently used in bio.tools, and the current development version (commit hash: 38f21c1edf839efa5d957395f9495562cc7dc1f8). The comparison of these two versions allows to foresee the impact of the future release of EDAM on bio.tools annotations.
The goal of this approach is mainly to provide a first assessment on the fitness EDAM and bio.tools, i.e. evaluate whether the space of scientific concepts available in EDAM is adapted to the semantic description of bio.tools entries, and highlight some curation priorities.
EDAM Concept categories
Each of the different EDAM sections analysed here are separated into the following categories:
-
Obsolete concepts: concepts that are judged to be obsolete in the current version. They are marked as deprecated but can still be referenced with their URI.
-
Auxiliary concepts: concepts which are usually placeholders to organize other sub-concepts and should not be used for annotation purposes. Technically speaking, these are identified using the "notRecommendedForAnnotation" property.
-
Valid concepts: concepts which are valid for annotation (neither deprecated, nor auxiliary).
-
New (valid) concepts: concepts added to the ontology in the current development version (future 1.26).
 {width=100%}
Figure 2: Usage of EDAM Topic concepts in bio.tools
{width=100%}
Figure 2: Usage of EDAM Topic concepts in bio.tools
As shown in Figure 2, EDAM topics usage in bio.tools reveals no major anomaly, most of the valid topics from version 1.25 being used in EDAM, whereas obsolete concepts are only marginally present in annotations. This could optimistically be interpreted as the indication of the fitness for the use of EDAM topics in bio.tools. However, this result doesn't guarantee that the topics section is extensive or precise enough, as some concepts can be used as default for lack of a better one.
 {width=100%}
Figure 3: Usage of EDAM Operation concepts in bio.tools
{width=100%}
Figure 3: Usage of EDAM Operation concepts in bio.tools
Figure 3 displays the same tendencies for EDAM operations, with an overall wide usage of valid operations while obsolete ones are mostly unused. However, a frequent issue is the use of auxiliary concepts. Although such annotations are probably accurate (i.e. consistent with the scientific function performed by the tool), the usage of such concepts is usually discouraged as they are either too broad or reserved for internal structural purposes. The status of some auxiliary concepts from EDAM might however be revised, if such concepts are the most relevant for annotation.
 {width=100%}
Figure 4: Usage of EDAM data concepts in bio.tools (only one plot is shown as no new data concepts have been added to the current development version of EDAM)
{width=100%}
Figure 4: Usage of EDAM data concepts in bio.tools (only one plot is shown as no new data concepts have been added to the current development version of EDAM)
 {width=100%}
Figure 5: Usage of EDAM format concepts in bio.tools
{width=100%}
Figure 5: Usage of EDAM format concepts in bio.tools
In contrast with the usage of topics and operations, a large proportion of data (23%) and formats (45.2%) are not used in bio.tools. Further investigation will be needed to determine this low usage is a consequence of annotation issues, or whether it is justified. Potential explanations include:
- a lower level of bio.tools EDAM annotation with data and formats, probably linked to the lack of data/format annotations in text-mining created entries.
- a scope difference between EDAM data and formats and bio.tools, as e.g. a number of EDAM formats might not be relevant in the scientific scope of bio.tools.
- the irrelevance of "legacy" formats related to life sciences technologies and tools which are now deprecated. It is important to note that such legacy file formats cannot be automatically deprecated as they are still used by older tools and can still be found in databases. This may explain the lack of usage of some valid formats in bio.tools.
Here, we sought to explore whether bio.tools identifiers, and by extension EDAM annotations, could be integrated with WorkflowHub entries for Galaxy workflows. WorkflowHub allows developers to register workflows, each of which are composed of one or more software tools. The connection to bio.tools is clear, and one can imagine a scenario where a workflow registered in WorkflowHub:
- Has component tools automatically extracted (as is the case currently for Galaxy);
- Each tool has a bio.tools identifier;
- This identifier allows WorkflowHub to import and present bio.tools annotations in workflow entries;
- WorkflowHub can filter workflows based on both EDAM terms and bio.tools identifiers (currently available functionality); and
- bio.tools can perform the reverse operation and import metadata about workflows that use specific bio.tools entries
To link WorkflowHub and bio.tools entries, all Galaxy workflows from WorkflowHub (https://workflowhub.eu/workflows) were accessed via API and, where metadata was available (82 out of 129 total workflows), a map was created between the individual tool steps in these workflow entries and Galaxy tool identifiers. This ultimately provided a list of workflow steps, mapped to identifiers from WorkflowHub, Galaxy and bio.tools.
| Metric | Value |
|---|---|
| No. of tools WITH a bio.tools ID | 513 |
| No. of tools without a bio.tools ID | 302 |
| Total no. of tools for all workflows | 815 |
| Total no. of workflows used | 82 |
The results of the mapping revealed that for 815 tools used across 82 workflows, 513 tools had a bio.tools identifier (63%). Note that the absence of a mapped bio.tools identifier does not mean that it does not exist. It is also possible that the identifier exists but that it still needs to be added to the Galaxy tool metadata. For example, hifiasm is used by the workflow PacBio HiFi genome assembly using hifiasm [@usesDataFrom:price_farquharson_2022]. This tool has a bio.tools identifier (https://bio.tools/hifiasm) which could be added to the Galaxy tool wrapper.
The table below shows the WorkflowHub identifier (with links) and the unique bio.tools identifiers extracted from 10 example workflows.
| WorkflowHub ID | bio.tools IDs |
|---|---|
| 138 | bedtools, bx-python, bcftools |
| 221 | hifiadapterfilt, bandage |
| 395 | cutadapt, bowtie2, samtools, bedtools, macs, multiqc |
| 397 | cutadapt, bowtie2, samtools, macs, multiqc |
| 398 | cutadapt, bowtie2, samtools, macs, multiqc |
| 399 | cutadapt, bowtie2, samtools, bedtools, seqcode, macs, multiqc |
| 400 | cutadapt, star, multiqc, cufflinks, bedtools |
| 403 | quast, busco, merqury |
| 406 | nanoplot, minimap2, Racon, unicycler, miniasm, bandage, staramr |
| 407 | bbmap, shovill, bwa, pilon, mob-suite, SISTR |
To facilitate the analysis of the data extracted from the Tools Ecosystem and other resources, we integrated them in a local SPARQL endpoint, using the GraphDB commercial software, a solution that enables the querying of RDF resources. The various resources uploaded to a GraphDB-based SPARQL endpoint include:
- the EDAM ontology [@usesDataFrom:jon_ison_2020_3899895], available in its development version on the EDAM GitHub repository.
- the bio.tools contents [@usesDataFrom:Ison2019], available on the Tools Platform Ecosystem git repository as a Turtle-formatted BioSchemas [@usesMethodIn:gray2017bioschemas] file.
- the workflow data extracted from WorkflowHub and Galaxy, and formatted as well in a BioSchemas format.
The analysis of the data is performed using SPARQL queries, which are performed using a number of Jupyter [@usesMethodIn:jupyter] notebooks. The various results are visualized using python libraries such as matplotlib [@usesMethodIn:hunter2007matplotlib].
The functions for mapping between WorkflowHub and bio.tools:
- Access the Galaxy Australia and Galaxy Europe APIs to extract both Galaxy specific tool identifiers and bio.tools identifiers, where available [@usesDataFrom:10.1093/nar/gkac247];
- Access the entire WorkflowHub registry via API (https://workflowhub.eu/workflows.json), filtering for Galaxy workflows only;
- Collect all available workflow metadata from the WorkflowHub API;
- For each workflow, extract all workflow step numbers and Galaxy identifiers (where these are documented, N = 82 of N = 129 workflows total);
- Map and extract Galaxy tool identifiers for each workflow step, WorkflowHub identifiers (i.e. unique number), bio.tools identifiers for each workflow step, and the workflow step numbers; and
- Convert mapped identifiers to
*.ttlformat
The functions described are available here. Code is based on previous work for ToolFinder [@usesMethodIn:gustafsson_ove_2021_5587837]. See also https://github.com/AustralianBioCommons/australianbiocommons.github.io.
The results presented here represent a first approach to building a knowledge base that integrates data from the various Tools Ecosystem components. Based on these results, we plan to develop further a series of goals, which we describe below:
-
Publication of an open SPARQL endpoint Improving on the results presented above, we will provide a publicly available SPARQL endpoint using an opensource software. This endpoint will be automatically updated by the Tools Ecosystem CI workflows. It will allow any user to query a dataset that will include not only EDAM, bio.tools, and WorkflowHub, but many other resources, and our teams of maintainers to run queries periodically if needed. It could also be used to optimize the Caseologue EDAM CI tool.
-
EDAM and Bio.tools analysis With this work we created and tested a valid work environment for analysing the EDAM ontology and the Bio.tools registry. In the same fashion as we plan on lifting up the POC level knowledge base up to an open public SPARQL endpoint, we plan on enriching the analyses to provide an exhaustive set of results that monitor the evolution and consistency of the Tools Ecosystem, for both maintainers and the Tools Ecosystem users. These will be run regularly by the Tools Ecosystem CI.
-
Thanks to our first query base we can easily imagine building on them to be able to evaluate and enhance metadata from the whole Ecosystem. Furthermore, we could to do so automatically to track progress using appropriate graphic visualizations, for the benefit of both maintainers and Tools Ecosystem communities and users.
This first round of analysis left us with some identified curation tasks for both EDAM and bio.tools that should be investigated in the future, as discussed in the Results and Discussion. Moreover, some potential enhancement were identified, such as the possibility to track the provenance of bio.tools entry annotations, which would allow to identify more easily e.g. whether they were generated by text-mining tools, or by human contributors.
This work also raises the question of the handling of deprecation in bio.tools. As of 2022, deprecated terms are not removed from the bio.tools annotation. This cannot be properly handled automatically and would be too time consuming manually. However, the total or partial automation of annotation updates could make use of "replacedBy" or "consider" EDAM properties which are used for deprecated concepts.
To resolve the lack of annotation of bio.tools entries using data and format concepts from EDAM, a perpective could be to improve bio.tools interface. All operations in EDAM are linked to a data via a "has_input" and a "has_output" relation (using inferences from parent concepts). For each operation added to the tool's annotation, input and output data would be suggested to the curator/author based on EDAM relation "has_input", "has_output". The same could go for suggestion of format based on its relation with data as 533 formats (over the 619 valid formats) are related to a data with the "is_format_of" relation. For human user we could also have in the bio.tools interface a "suggested input/output" that would be displayed on the tool page but clearly identifed as an unverified annotation. The enhancement of complete EDAM annotation could lead to automatic workflow generation using the whole Ecosystem metadata.
In the previous section we detailed new approaches for evaluating and improving EDAM annotations in bio.tools: the other side of this coin is to ensure that other parts of the Ecosystem can share and benefit from these improvements. One of these parts is computational workflows that are registered in WorkflowHub. Each of these workflows represent a significant investment of researcher time and expertise: ideally these workflows would be able to draw directly on the wealth of metadata and EDAM ontology annotations stored by a registry like bio.tools, with minimal additional input of effort from a workflow developer. The prototype map described by this project is incomplete, with 37% of Galaxy workflow tools missing a bio.tools identifier. Likely causes include either missing annotations in the Galaxy tool wrappers or missing bio.tools registry entries. However, the potential value is already clear: a potential next step is for the map functions between WorkflowHub and Galaxy to be productionised by the Tools Ecosystem, such that bio.tools is able to access all Galaxy workflows on WorkflowHub (N = 129 in January 2023) that make use of bio.tools entries, and WorkflowHub is able to access tool components of its workflows as well as bio.tools registry metadata. Synchronisation in this manner will give each platform the opportunity to further improve the experience of users that contribute to and maintain a FAIR software ecosystem.
Many thousands of Galaxy workflows exist globally. With automated integration, users of WorkflowHub will be able to intuitively navigate the growing set of Galaxy workflows based on their tool of choice, topic, or software operation.
The code described to run the analyses and obtain the results presented here is freely available on a dedicated GitHub repository. The data collected are also freely available on the Tools Ecosystem main repository and on the EDAM repository.
This work was funded/supported by ELIXIR, the research infrastructure for life-science data. This work was supported by the Australian BioCommons which is enabled by NCRIS via Bioplatforms Australia funding. The authors wish to thank the WorkflowHub, Bioschemas, and Galaxy project teams for their technical help and the fruitful discussions that led to these results.