- Quick start
- Requirements
- Installation
- Full pipeline documentation
- Case studies
- PBMC 10k dataset (10x Genomics)
- Full SCENIC analysis, plus filtering, clustering, visualization, and SCope-ready loom file creation:
- Extended analysis post-SCENIC:
- Cancer data sets
- Mouse brain data set
- PBMC 10k dataset (10x Genomics)
- References and more information
We recommend using this notebook as a template for running an interactive analysis in Jupyter. See the installation instructions for information on setting up a kernel with pySCENIC and other required packages.
The following tools are required to run the steps in this Nextflow pipeline:
- Nextflow
- A container system, either of:
The following container images will be pulled by nextflow as needed:
- Docker: aertslab/pyscenic:latest.
- Singularity: aertslab/pySCENIC:latest.
- See also here.
Download a minimum set of SCENIC database files for a human dataset (approximately 78 MB). This small test dataset takes approximately 30s to run using 6 threads on a standard desktop computer.
mkdir example && cd example/
# Transcription factors:
wget https://raw.githubusercontent.com/aertslab/SCENICprotocol/master/example/allTFs_hg38.txt
# Motif to TF annotation database:
wget https://raw.githubusercontent.com/aertslab/SCENICprotocol/master/example/motifs.tbl
# Ranking databases:
wget https://raw.githubusercontent.com/aertslab/SCENICprotocol/master/example/genome-ranking.feather
# Finally, get a small sample expression matrix (loom format):
wget https://raw.githubusercontent.com/aertslab/SCENICprotocol/master/example/expr_mat.loom
Either Docker or Singularity images can be used by specifying the appropriate profile (-profile docker or -profile singularity).
nextflow run aertslab/SCENICprotocol \
-profile docker \
--loom_input expr_mat.loom \
--loom_output pyscenic_integrated-output.loom \
--TFs allTFs_hg38.txt \
--motifs motifs.tbl \
--db *feather
By default, this pipeline uses the container tag specified by the --pyscenic_tag parameter.
This is currently set to 0.9.16, which uses a container with both pySCENIC and Scanpy 1.4.4.post1 installed.
A custom container can be used (e.g. one built on a local machine) by passing the name of this container to the --pyscenic_container parameter.
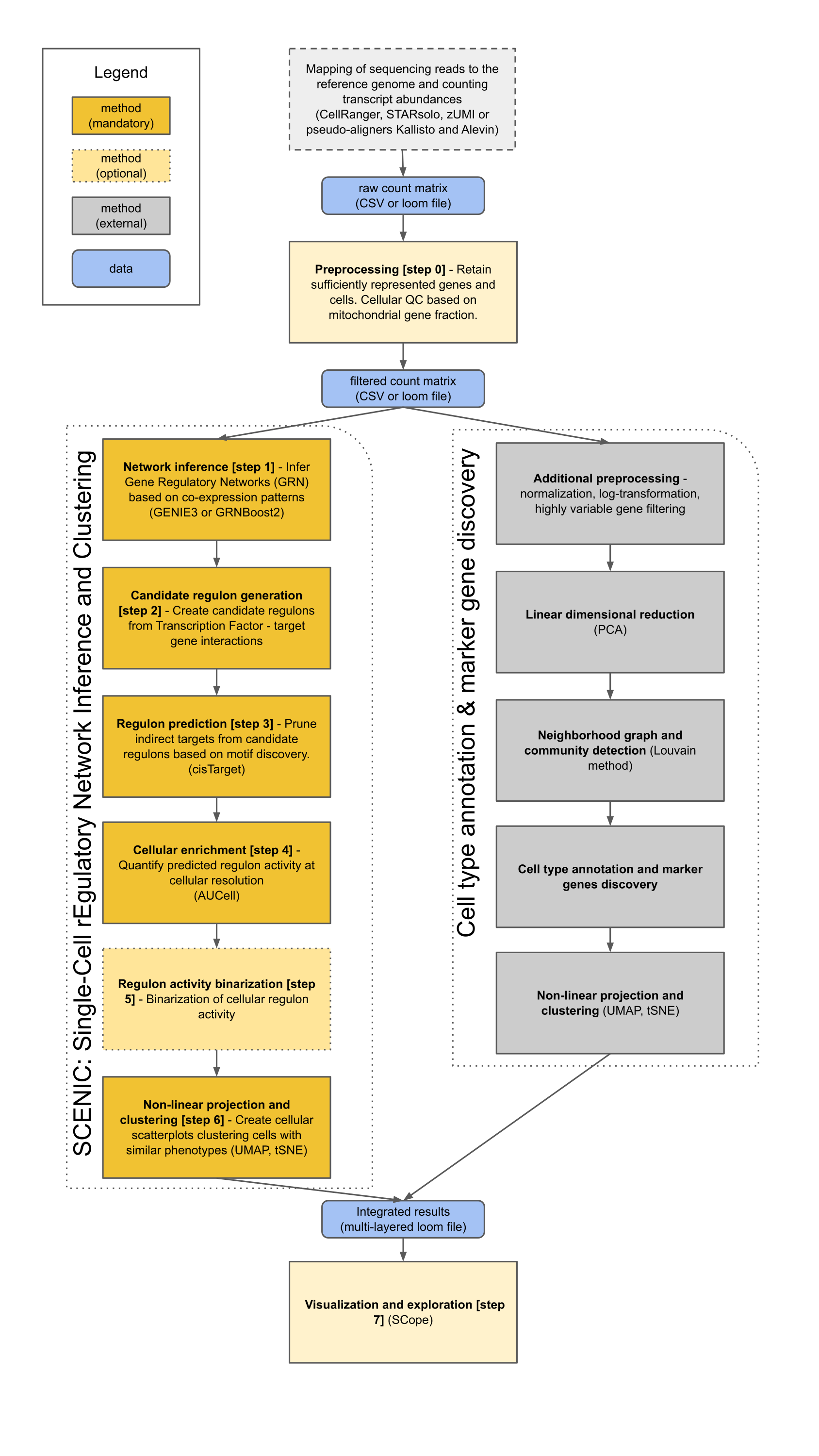
The output of this pipeline is a loom-formatted file (by default: output/pyscenic_integrated-output.loom) containing:
* The original expression matrix
* The pySCENIC-specific results:
* Regulons (TFs and their target genes)
* AUCell matrix (cell enrichment scores for each regulon)
* Dimensionality reduction embeddings based on the AUCell matrix (t-SNE, UMAP)
* Results from the parallel best-practices analysis using highly variable genes:
* Dimensionality reduction embeddings (t-SNE, UMAP)
* Louvain clustering annotations
- Python version 3.6 or greater
- Tested on various Unix/Linux distributions (Ubuntu 18.04, CentOS 7.6.1810, MacOS 10.14.5)