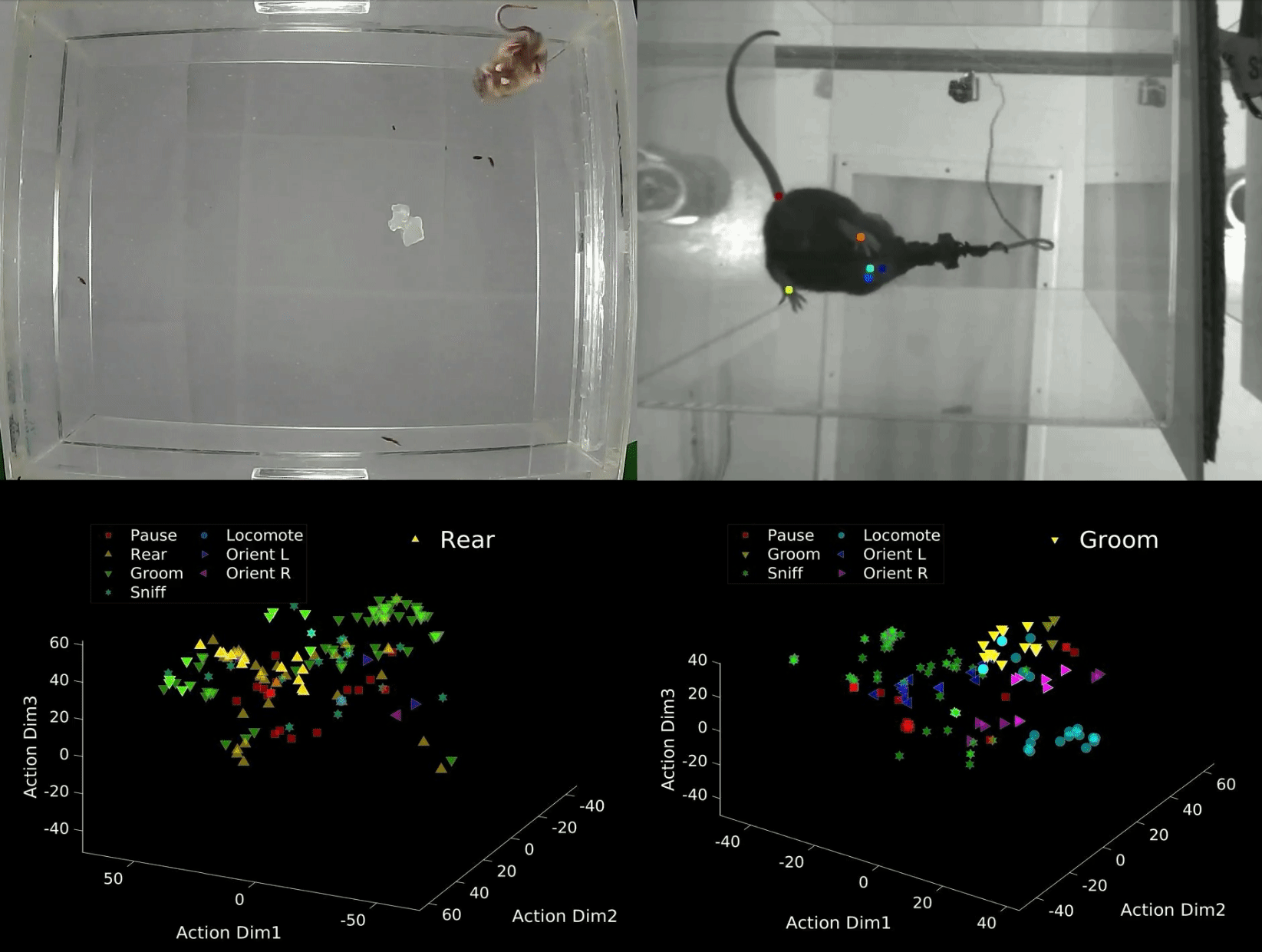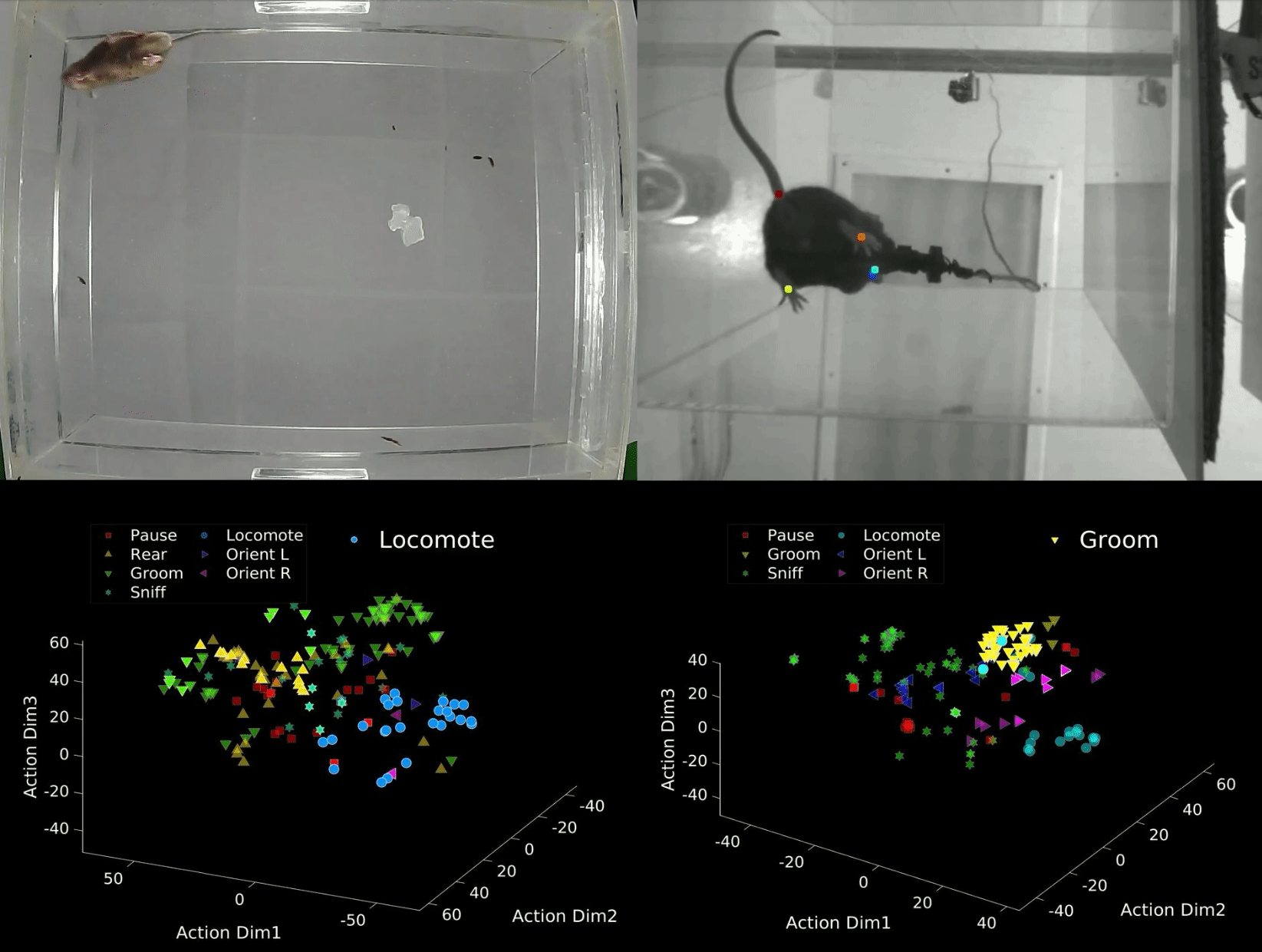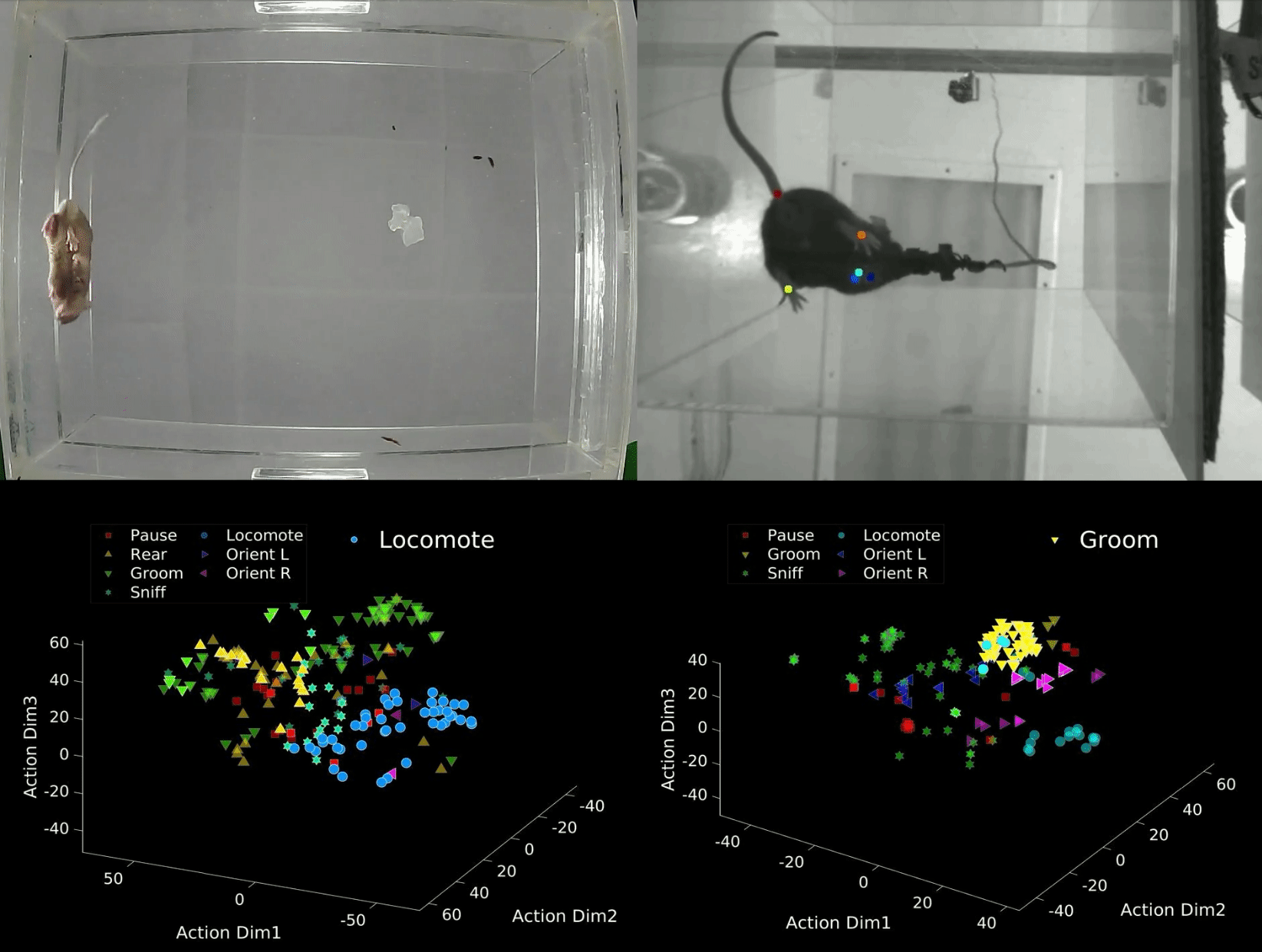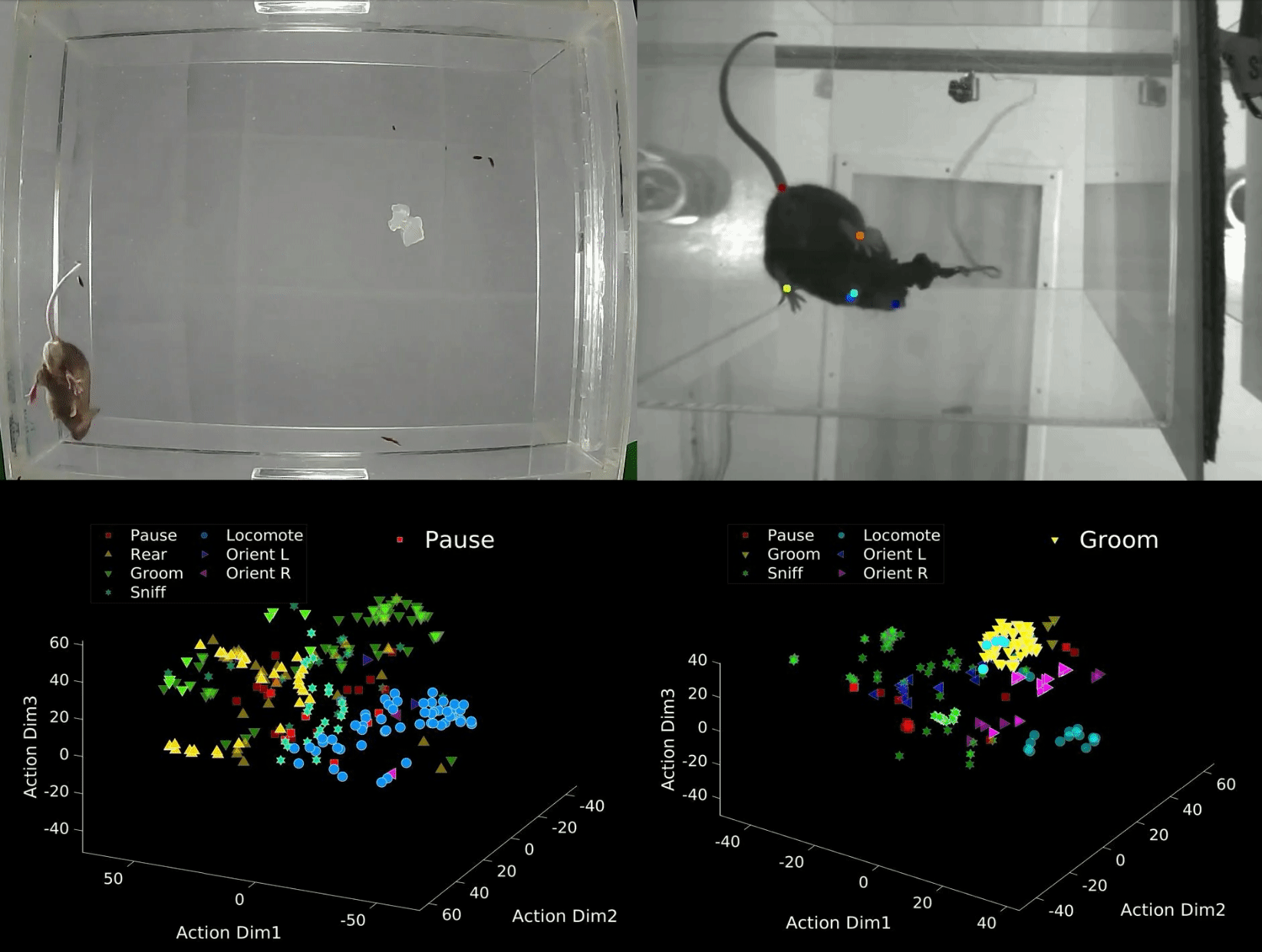
B-SOID ("B-side") is an unsupervised learning algorithm written in MATLAB that analyzes sub-second rodent behavior from a single bottom-up recording video-camera. With the output trajectory of 6 body parts (snout, the 4 paws, and the base of the tial) of a rodent extracted from DeepLabCut1,2, this algorithm performs t-Distributed Stochastic Neighbor Embedding (t-SNE4, MATLAB©) of the 7 heirarchically different time-varying signals to fit Gaussian Mixture Models5. The output agnostically separates statistically significant distributions in the 3-dimensional action space and are found to be correlated with different observable rodent behaviors. Although the primary purpose of this integrated technology is for automatic classification of behaviors that either have rodent-to-rodent variability, or carry observer bias, the output also uncovers behaviors likely coincides better with latent variables. The idea behind this also is also expandable to multiple perspectives and other fields of study.
Use Git using the web URL or download ZIP.
Change your current working directory to the location where you want the cloned directory to be made.
git clone https://github.com/YttriLab/B-SOID.gitChange MATLAB current folder to B-SOID/bsoid
Import .csv file, and convert it to a matrix Using the demo mouse navigating the open-field from the Yttri-Lab
data_struct = import(Ms2OpenField.csv);
rawdata = data_struct.dataApply a low-pass filter for data likelihood. dlc_preprocess finds the most recent x,y that are above the threshold and replaces with them. Refer to dlc_preprocess.md.
Based on our pixel-error, the Yttri-Lab decided to go with 0.5 as the likelihood threshold.
data = dlc_preprocess(rawdata,0.5); Option 1: Manual criteria for a rough but fast analyses (If you are interested in considering the rough estimate of the 7 behaviors: 1 = Pause, 2 = Rear, 3 = Groom, 4 = Sniff, 5 = Locomote, 6 = Orient Left, 7 = Orient Right). Refer to bsoid_mt.md
Based on our zoom from the 15 inch x 12 inch open field set-up, at a camera resolution of 1280p x 720p, the Yttri-Lab has set criteria for the 7 states of action. This fast algorithm was able to pull out proof-of-concept Parkisonian mouse in the Yttri-Lab. This can also be a first pass at analyzing biases in transition matrices, as well as overarching behavioral changes before digging further into the behavior.
[g_label,g_num,perc_unk] = bsoid_fast(data,pix_cm); % data, pixel/cm Option 2: Unsupervised grouping of action space, more refined and reliable output (This can uncover behaviors, not just the 7 listed above, and perhaps coincide better with latent variables).
- The following steps are only valid if you go with
Option 2
Unsupervised grouping of the purely data-driven action space. Refer to bsoid_gmm.md
Based on the comparable results benchmarked against human observers for the Yttri-Lab dataset, we also tested the generalizability with a dataset from the Ahmari Lab and found that the agnostic data-driven approach allowed for scaling to the zoom as well as animal-animal variability. It will also sub-divide what seems to be the same action groups into different categories, of which may or may not be important depending on the study.
[f,tsne_f,grp,llh,bsoid_fig] = bsoid_gmm(data,60); % data, sampling-rateDefine threshold for the 7 actions (i.e. if majority of data in that unsupervised grouping has a angular change, it is turning)
(OPTIONAL) Step VI (If you are interested in creating short videos (.avi) of the groups). This is not recommended if it has more than 100,000 frames at 7201280p.*
vidObj = VideoReader(filenamevid); % video used to generate DLCAssuming all behaviors can be sampled from the first 10 minutes (600 seconds), have MATLAB store only every 10 frames per second.
k = 1; kk = 1;
while vidObj.CurrentTime < 599.9
vidObj.CurrentTime = k/60;
video{i}(kk).cdata = readFrame(vidObj); % save only every 10fps
kk = kk+1; % save only every 10fps
k = k+round(vidObj.FrameRate)/10; % reduce down to 10fps (100ms/frm)
endCreate short videos in the desired output folder (default = current directory) of different groups of action clusters that at least lasted for ~300ms, and slow the video down to 0.75X for better understanding.
filepathout = uigetdir;
[t,b,b_ex] = action_gif(video,grp_fill,1,5,6,0.75,filepathout);Pull requests are welcome. For recommended changes that you would like to see, open an issue.
We are a neuroscience lab and can easily foresee this as being upgraded. Please do not hesitate in helping us working towards a more efficient and accurate algorithm. All major contributors will be cited in our publications.
This software package provided without warranty of any kind and is licensed under the GNU Lesser General Public License v3.0. Cite us if you use the code and/or data!.https://choosealicense.com/licenses/mit/)