GADMA was updated and tested on Python3.10 with the latest versions of dependencies.
Changelog since 2.0.0:
- Add lower bounds for first and second splits (see #92)
- Create pyproject.toml and setup.cfg
- Add dependencies including moments-popgen, demes and demesdraw to setup - they will be automatically installed.
- Change code to work with last version of moments.
- Drop support for Gpy and GPyOpt bayesian optimization
- Tests for SMAC bayesian optimization does not run on GitHub Actions - need to upgrade code for the last smac version (TODO).
- Update params_file template
- Update docs
Major release Official release of GADMA2.0.0, it includes all pre-releases 2.0.0rc1 - 2.0.0rc26
Changelog since 2.0.0rc26:
- Catch cases when theta and ancestral population size are negative for moments and dadi engines (issue #84). Fix optimization routine to ignore such parameters.
- Fix GitHub Action tests and PyPi publishing.
- Add documentation (regarding issues #60, #75, #85)
- Add note when SFS is build from less than 90% of given SNPs.
- Option Split fraction is set to be False by default.
- Update code according with new version of moments.LD that has nus in steady_state.
- Fix float problem with new versions of numpy.
- Fix bug with demes: it multiplied time twice by the generation time.
- Fix CLAIC bug
- Fix bug with momentsLD engine for models with exponential or linear change (incorrect likelihood).
- Restrict momi2 to use only one core.
- Use another scipy function for Nelder_mead algorithm (local)
- Make all histories with different parameters when structure is changed (e.g. from 1,1 to 2,1)
- Allow usage of models specified using gadma API, but no documentation on that.
- Fix bug when failed model is reported as the best.
- Add additional checks for Bayesian optimization
- Add documentation about BO
- Linear extrapolation for dadi was integrated through
Dadi extrapolationoption. - Engine momi is renamed in momi2.
- Inference of dominance rates was added in GADMA interface.
- Fix bug with CLAIC evaluation (for any eps it was taken as 1e-2).
- Hyperparameters of genetic algorithm were updated.
- Fix minor bugs.
- Fix documentation
- Genetic algorithm is allowed to be run after Bayesian optimization with --resume option.
- Momi engine can read .gz files.
- Minor bug fixes.
Major release
- We present a new engine -
momentsLD! It is the first engine that use LD statistics for demographic inference in GADMA. For more information please see documentation. As in case of themomiengine if there are some problems with usage ofmomentsLDengine please post an issue on GitHub, we really appreciate the feedback! - Several bugs were fixed including Issue #58.
Major release
- We present a new engine -
momi! It is now possible to perform demographic inference withmomiengine as well as draw pictures of models with it. For more information please see documentation. If one will face problems with usage ofmomiengine please post an issue on GitHub, we really appreciate the feedback! - Option
Recombination ratewas added as future engines integrated into GADMA can reqiure it. ALsomomineeds it to simulate data withmsprime. - New input data - fastsimcoal2 input files. All three engines (
dadi,momentsandmomi) are able to read this format now. For more information about format please see corresponding section in documentation. - GADMA is now available via
conda(Bioconda). - We fix an error in a genetic algorithm that had ignored change of mutation rate and strength of GA. Unfortunately, the hyperparameter optimization that was performed before is no longer valid. We are going to rerun and obtain new hyperparameter values as soon as possible. Right now we change values of mutation rate and strength constants to default values.
- The distributions of variable sampling were also updated: the time variable will be sampled from log normal distribution (was just normal) and option
Random NAis False by default now. According to the experiments on several datasets GADMA with new distributions performs better.
- VCF data format was add as one of input data for GADMA! Now SFS data can be build from VCF and popmap files like:
# param_file
Input data: vcf_file, popmap_file
Input filesetting changed toInput dataoption.
- Add Bayesian optimizations to GADMA. There are three versions of it:
- GPyOpt_Bayesian_optimization
- SMAC_squirrel_optimization
- SMAC_BO_optimization
- Add
demesas a new engine for models plotting. - Add
Inbreedingto infer inbreeding coefficients withdadiengine. - Update interface of
gadma.optimizers. - Move
Multinomoption from deprecated options to changed. AddAncestral size as parameterinsteadMultinom. - Update docs.
Local optimizations got hyperparameters from dadi and moments. Now they work and are efficient the same way as in those packages.
Prerelease of GADMA v2.0.0.
Code of GADMA was updated in order to make it more stable and accurate. There are tests for implementation and online documentation on ReadTheDocs.
GADMA is now available via pip and has better optimization algorithm!
Updated perparameters of genetic algorithm
We have tuned hyperparameters of the genetic algorithm by Bayesian optimization implemented in SMAC software. The following hyperparameters were optimized:
| Hyperparameter | Old value | New value |
|---|---|---|
| Mean mutation rate | 0.2 | 0.453272 |
| Const_for_mutation_rate | 1.2 | 1.068062 |
| Mean mutation strength | 0.2 | 0.625049 |
| Const for mutation strength | 1.1 | 1.016571 |
| Fraction of mutated individuals | 0.3 | 0.55560528752 |
| Fraction of crossed individuals | 0.3 | 0.18828153004 |
| Fraction of random generated individuals | 0.2 | 0.12600048532 |
Four different combinations of hyperparameters were optimized with SMAC. This 4th combination provided the best performance on train and test data.
SMAC was launched for 10,000 iterations in 10 parallel runs for 14 days. Four datasets (instances) were used as training data for optimization. We allowed maximum of 50 runs on each of train instances.
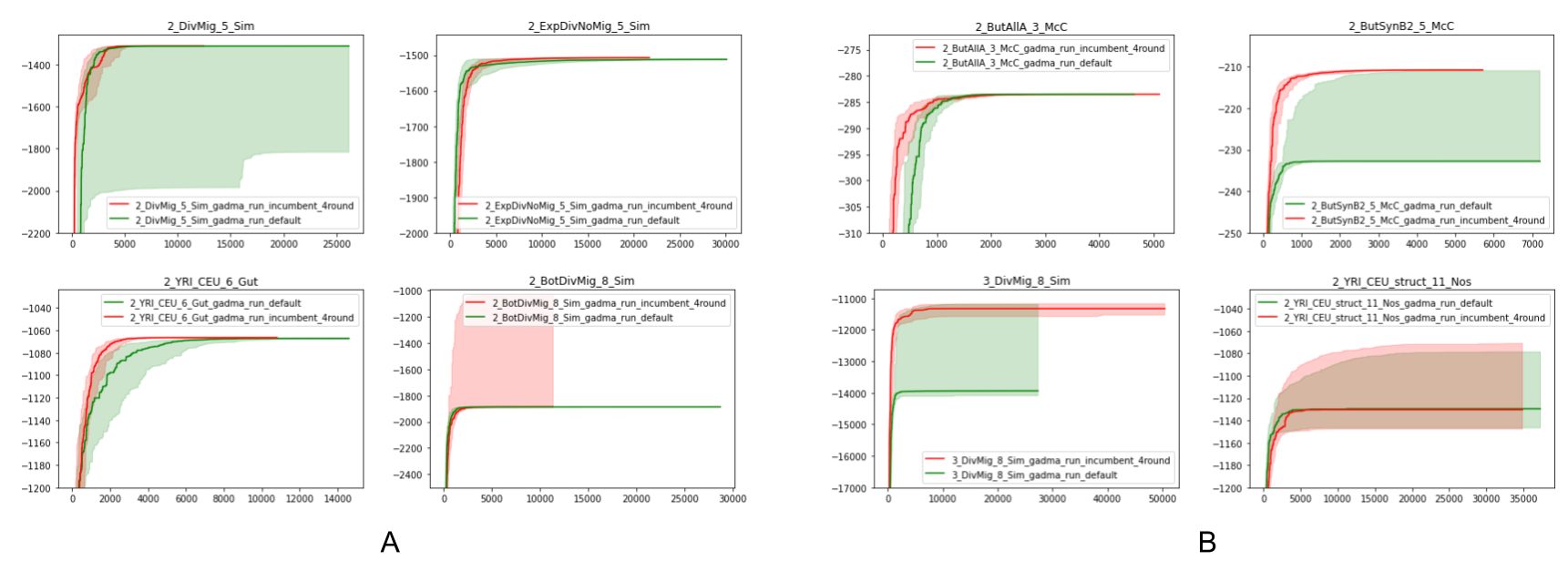
Picture above shows the comparison of genetic algorithms with different values of hyperparameters on train and test datasets. Green color corresponds to GADMA v1 and red color for GADMA v2. The abscissa axis presents iterations (log-likelihood evaluations), the ordinate refers to the value of log-likelihood. Colored lines correspond to the medians of best log-likelihoods values (50 runs) and shadowed areas are ranges between first (0.25) and third (0.75) quartiles. (A) Convergence on train datasets (B) Convergence on test datasets.
Updated options names in parameters file
Some options in parameters file were changed. Some of them have new names:
Use moments or dadi->Engine,Size of population in ga->Size of generation,Fractions in ga->Fractions,Epsilon->Eps,Stop iteration->Stuck generation number,Name of local optimization->Local optimizer,Lower bounds->Lower bound,Upper bounds->Upper bound,Verboseoption is now both for the genetic algorithm and the local search as verbosity of the output.
It is still possible to use old names - GADMA will successfully read it and print the following warning:
UserWarning: Setting `Use moments or dadi` is renamed in 2 version of GADMA to `Engine`. It is successfully read. (/home/build/ctlab/GADMA/gadma/cli/settings_storage.py:741
Deprecated options names in parameters file
Some options are deprecated:
multinom,flush_delay,epsilon_for_ls,gtol,maxiter,multinomial_mutation,multinomial_crossing,distribution,std,mean_mutation_rate_for_hc,const_for_mutation_rate_for_hc,stop_iteration_for_hc.In general those options were in extra parameters file as options of local search algorithms and hill climbing. Hill climbing algorithm if now fully deprecated.
GADMA prints the following warning if some of deprecated options are set in parameters file:
UserWarning: Setting `Multinom` was deprecated in 2 version of GADMA. If you have not set it in purpose, ignore this warning. (/home/build/ctlab/GADMA/gadma/cli/settings_storage.py:747)
New options for mutation rate and sequence length
Option
Theta0is required to translate parameters from genetic units.Theta0is mutation flux equal to4 mu L, wheremu- mutation rate per base per generation andL- length of sequence. Now it is possible to set mutation rate and sequence length instead of theta0:
Mutation rate- mutation rate per base per generation.Sequence length- length of sequence that was used to build data.
New options for migrations
New options for migrations. Now migrations could be symmetrical, also some of them could be restricted manually:
Symmetric migrations- ifTruethen all migrations are symmetrical.Migration masks- masks for migration matrices for all time intervals with migrations. Consists of 0 and 1, where 0 means that migration is missed and equal to zero.
Other new options
Outgroup- new option for data. IfTruethen data has outgroup and AFS is not folded.Split fractions- ifTruethen population is divided into two new according to the fraction which determines the sizes. In such case sum of newly formed populations is equal to size of parent population. If option isFalsethen each newly formed population has its own independent size.Vmin- minimal value to draw on heatmap of the AFS data. Is useful when pictures are not good.Some additional options for the genetic algorithm (it is equivalent of the
Fractions):
n_elitism- number of solutions to take to the new generation.p_mutation- probability of mutated solution in the new generation.p_crossover- probability of crossover solution in the new generationp_random- probability of random generated solution in the new generation.
New local search algorithms
Now GADMA has full set of local search method for any engine. Hill climbing is deprecated. Other algorithms have new names in additional to those from
dadi/moments. One could call by one of two names and it will be the same algorithm.
- L-BFGS-B algorithm is available by names
L-BFGS-B,optimize_lbfgsbandL-BFGS-B_log,optimize_log_lbfgsbto apply logarithm to search space.- BFGS is available under
BFGS,optimizeandBFGS_log,optimize_log.- Powell's method is available under
Powell,optimize_powellandPowell_log,optimize_log_powell.- Nelder-Mead algorithm is available under
Nelder-Mead,optimize_fminandNelder-Mead_log,optimize_log_fmin.- No local optimization is available under name
None.
New examples
API