| title | subtitle | author | job | logo | framework | highlighter | hitheme | url | widgets | mode | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Classes and Methods in R |
Roger D. Peng, Associate Professor of Biostatistics |
Johns Hopkins Bloomberg School of Public Health |
bloomberg_shield.png |
io2012 |
highlight.js |
tomorrow |
|
|
selfcontained |
-
A system for doing object oriented programming
-
R was originally quite interesting because it is both interactive and has a system for object orientation.
- Other languages which support OOP (C++, Java, Lisp, Python, Perl) generally speaking are not interactive languages
-
In R much of the code for supporting classes/methods is written by John Chambers himself (the creator of the original S language) and documented in the book Programming with Data: A Guide to the S Language
-
A natural extension of Chambers’ idea of allowing someone to cross the user −→ programmer spectrum
-
Object oriented programming is a bit different in R than it is in most languages — even if you are familiar with the idea, you may want to pay attention to the details
S3 classes/methods
- Included with version 3 of the S language.
- Informal, a little kludgey
- Sometimes called old-style classes/methods
S4 classes/methods
- more formal and rigorous
- Included with S-PLUS 6 and R 1.4.0 (December 2001)
- Also called new-style classes/methods
- For now (and the forseeable future), S3 classes/methods and S4 classes/methods are separate systems (but they can be mixed to some degree).
- Each system can be used fairly independently of the other.
- Developers of new projects (you!) are encouraged to use the S4 style classes/methods.
- Used extensively in the Bioconductor project
- But many developers still use S3 classes/methods because they are “quick and dirty” (and easier).
- In this lecture we will focus primarily on S4 classes/methods
- The code for implementing S4 classes/methods in R is in the methods package, which is usually loaded by default (but you can load it with
library(methods)if for some reason it is not loaded)
- A class is a description of an thing. A class can be defined using
setClass()in the methods package. - An object is an instance of a class. Objects can be created using
new(). - A method is a function that only operates on a certain class of objects.
- A generic function is an R function which dispatches methods. A generic function typically encapsulates a “generic” concept (e.g.
plot,mean,predict, ...)- The generic function does not actually do any computation.
- A method is the implementation of a generic function for an object of a particular class.
- The help files for the ‘methods’ package are extensive — do read them as they are the primary documentation
- You may want to start with
?Classesand?Methods - Check out
?setClass,?setMethod, and?setGeneric - Some of it gets technical, but try your best for now—it will make sense in the future as you keep using it.
- Most of the documentation in the methods package is oriented towards developers/programmers as these are the primary people using classes/methods
All objects in R have a class which can be determined by the class function
class(1)## [1] "numeric"
class(TRUE)## [1] "logical"
class(rnorm(100))## [1] "numeric"
class(NA)## [1] "logical"
class("foo")## [1] "character"
Data classes go beyond the atomic classes
x <- rnorm(100)
y <- x + rnorm(100)
fit <- lm(y ~ x) ## linear regression model
class(fit)## [1] "lm"
- S4 and S3 style generic functions look different but conceptually, they are the same (they play the same role).
- When you program you can write new methods for an existing generic OR create your own generics and associated methods.
- Of course, if a data type does not exist in R that matches your needs, you can always define a new class along with generics/methods that go with it
The mean and print functions are generic
mean## function (x, ...)
## UseMethod("mean")
## <bytecode: 0x7facdb660ad0>
## <environment: namespace:base>
print## function (x, ...)
## UseMethod("print")
## <bytecode: 0x7facd9ccfd58>
## <environment: namespace:base>
The mean generic function has a number of methods associated with it.
methods("mean")## [1] mean.Date mean.default mean.difftime mean.POSIXct mean.POSIXlt
The show function is from the methods package and is the S4
equivalent of print
show## standardGeneric for "show" defined from package "methods"
##
## function (object)
## standardGeneric("show")
## <bytecode: 0x7facdb8034d8>
## <environment: 0x7facdb779868>
## Methods may be defined for arguments: object
## Use showMethods("show") for currently available ones.
## (This generic function excludes non-simple inheritance; see ?setIs)
The show function is usually not called directly (much like print)
because objects are auto-printed.
showMethods("show")## Function: show (package methods)
## object="ANY"
## object="classGeneratorFunction"
## object="classRepresentation"
## object="envRefClass"
## object="function"
## (inherited from: object="ANY")
## object="genericFunction"
## object="genericFunctionWithTrace"
## object="MethodDefinition"
## object="MethodDefinitionWithTrace"
## object="MethodSelectionReport"
## object="MethodWithNext"
## object="MethodWithNextWithTrace"
## object="namedList"
## object="ObjectsWithPackage"
## object="oldClass"
## object="refClassRepresentation"
## object="refMethodDef"
## object="refObjectGenerator"
## object="signature"
## object="sourceEnvironment"
## object="standardGeneric"
## (inherited from: object="genericFunction")
## object="traceable"
The first argument of a generic function is an object of a particular class (there may be other arguments)
- The generic function checks the class of the object.
- A search is done to see if there is an appropriate method for that class.
- If there exists a method for that class, then that method is called on the object and we’re done.
- If a method for that class does not exist, a search is done to see if there is a default method for the generic. If a default exists, then the default method is called.
- If a default method doesn’t exist, then an error is thrown.
- You cannot just print the code for a method like other functions because the code for the method is usually hidden.
- If you want to see the code for an S3 method, you can use the function
getS3method. - The call is
getS3method(<generic>, <class>) - For S4 methods you can use the function
getMethod - The call is
getMethod(<generic>, <signature>)(more details later)
What’s happening here?
set.seed(2)
x <- rnorm(100)
mean(x)## [1] -0.0307
- The class of x is “numeric”
- But there is no mean method for “numeric” objects!
- So we call the default function for
mean.
head(getS3method("mean", "default"), 10)##
## 1 function (x, trim = 0, na.rm = FALSE, ...)
## 2 {
## 3 if (!is.numeric(x) && !is.complex(x) && !is.logical(x)) {
## 4 warning("argument is not numeric or logical: returning NA")
## 5 return(NA_real_)
## 6 }
## 7 if (na.rm)
## 8 x <- x[!is.na(x)]
## 9 if (!is.numeric(trim) || length(trim) != 1L)
## 10 stop("'trim' must be numeric of length one")
tail(getS3method("mean", "default"), 10)##
## 15 if (any(is.na(x)))
## 16 return(NA_real_)
## 17 if (trim >= 0.5)
## 18 return(stats::median(x, na.rm = FALSE))
## 19 lo <- floor(n * trim) + 1
## 20 hi <- n + 1 - lo
## 21 x <- sort.int(x, partial = unique(c(lo, hi)))[lo:hi]
## 22 }
## 23 .Internal(mean(x))
## 24 }
What happens here?
set.seed(3)
df <- data.frame(x = rnorm(100), y = 1:100)
sapply(df, mean)## x y
## 0.01104 50.50000
-
The class of
dfis "data.frame"; each column can be an object of a different class -
We
sapplyover the columns and call themeanfunction -
In each column,
meanchecks the class of the object and dispatches the appropriate method. -
We have a
numericcolumn and anintegercolumn;meancalls the default method for both
-
Some S3 methods are visible to the user (i.e.
mean.default), -
Never call methods directly
-
Use the generic function and let the method be dispatched automatically.
-
With S4 methods you cannot call them directly at all
The plot function is generic and its behavior depends on the object being plotted.
set.seed(10)
x <- rnorm(100)
plot(x)For time series objects, plot connects the dots
set.seed(10)
x <- rnorm(100)
x <- as.ts(x) ## Convert to a time series object
plot(x)If you write new methods for new classes, you’ll probably end up writing methods for the following generics:
- print/show
- summary
- plot
There are two ways that you can extend the R system via classes/methods
- Write a method for a new class but for an existing generic function (i.e. like
print) - Write new generic functions and new methods for those generics
Why would you want to create a new class?
- To represent new types of data (e.g. gene expression, space-time, hierarchical, sparse matrices)
- New concepts/ideas that haven’t been thought of yet (e.g. a fitted point process model, mixed-effects model, a sparse matrix)
- To abstract/hide implementation details from the user I say things are “new” meaning that R does not know about them (not that they are new to the statistical community).
A new class can be defined using the setClass function
- At a minimum you need to specify the name of the class
- You can also specify data elements that are called slots
- You can then define methods for the class with the
setMethodfunction Information about a class definition can be obtained with theshowClassfunction
Creating new classes/methods is usually not something done at the console; you likely want to save the code in a separate file
library(methods)
setClass("polygon",
representation(x = "numeric",
y = "numeric"))-
The slots for this class are
xandy -
The slots for an S4 object can be accessed with the
@operator.
A plot method can be created with the setMethod function.
-
For
setMethodyou need to specify a generic function (plot), and a signature. -
A signature is a character vector indicating the classes of objects that are accepted by the method.
-
In this case, the
plotmethod will take one type of object, apolygonobject.
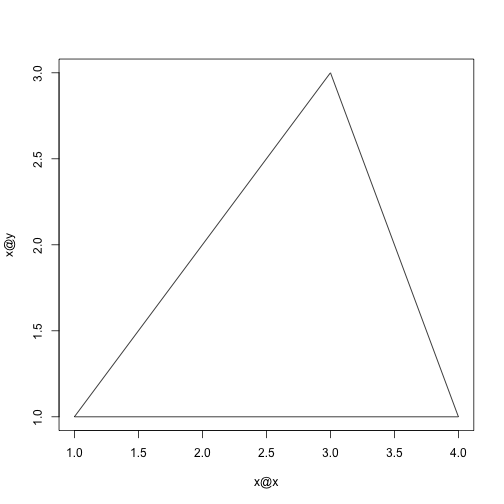
Creating a plot method with setMethod.
setMethod("plot", "polygon",
function(x, y, ...) {
plot(x@x, x@y, type = "n", ...)
xp <- c(x@x, x@x[1])
yp <- c(x@y, x@y[1])
lines(xp, yp)
})## Creating a generic function for 'plot' from package 'graphics' in the global environment
## [1] "plot"
- Notice that the slots of the polygon (the x- and y-coordinates) are
accessed with the
@operator.
After calling setMethod the new plot method will be added to the list of methods for plot.
library(methods)
showMethods("plot")## Function: plot (package graphics)
## x="ANY"
## x="polygon"
Notice that the signature for class polygon is listed. The method for ANY is the default method and it is what is called when now other signature matches
p <- new("polygon", x = c(1, 2, 3, 4), y = c(1, 2, 3, 1))
plot(p)-
Developing classes and associated methods is a powerful way to extend the functionality of R
-
Classes define new data types
-
Methods extend generic functions to specificy the behavior of generic functions on new classes
-
As new data types and concepts are created, classes/methods provide a way for you to develop an intuitive interface to those data/concepts for users
-
The best way to learn this stuff is to look at examples
-
There are quite a few examples on CRAN which use S4 classes/methods. You can usually tell if they use S4 classes/methods if the methods package is listed in the
Depends:field -
Bioconductor (http://www.bioconductor.org) — a rich resource, even if you know nothing about bioinformatics
-
Some packages on CRAN (as far as I know) — SparseM, gpclib, flexmix, its, lme4, orientlib, filehash
-
The
stats4package (comes with R) has a bunch of classes/methods for doing maximum likelihood analysis.