Learning Implicit Surfaces from Point Clouds
This is our implementation of Points2Surf, a network that estimates a signed distance function from point clouds. This SDF is turned into a mesh with Marching Cubes.
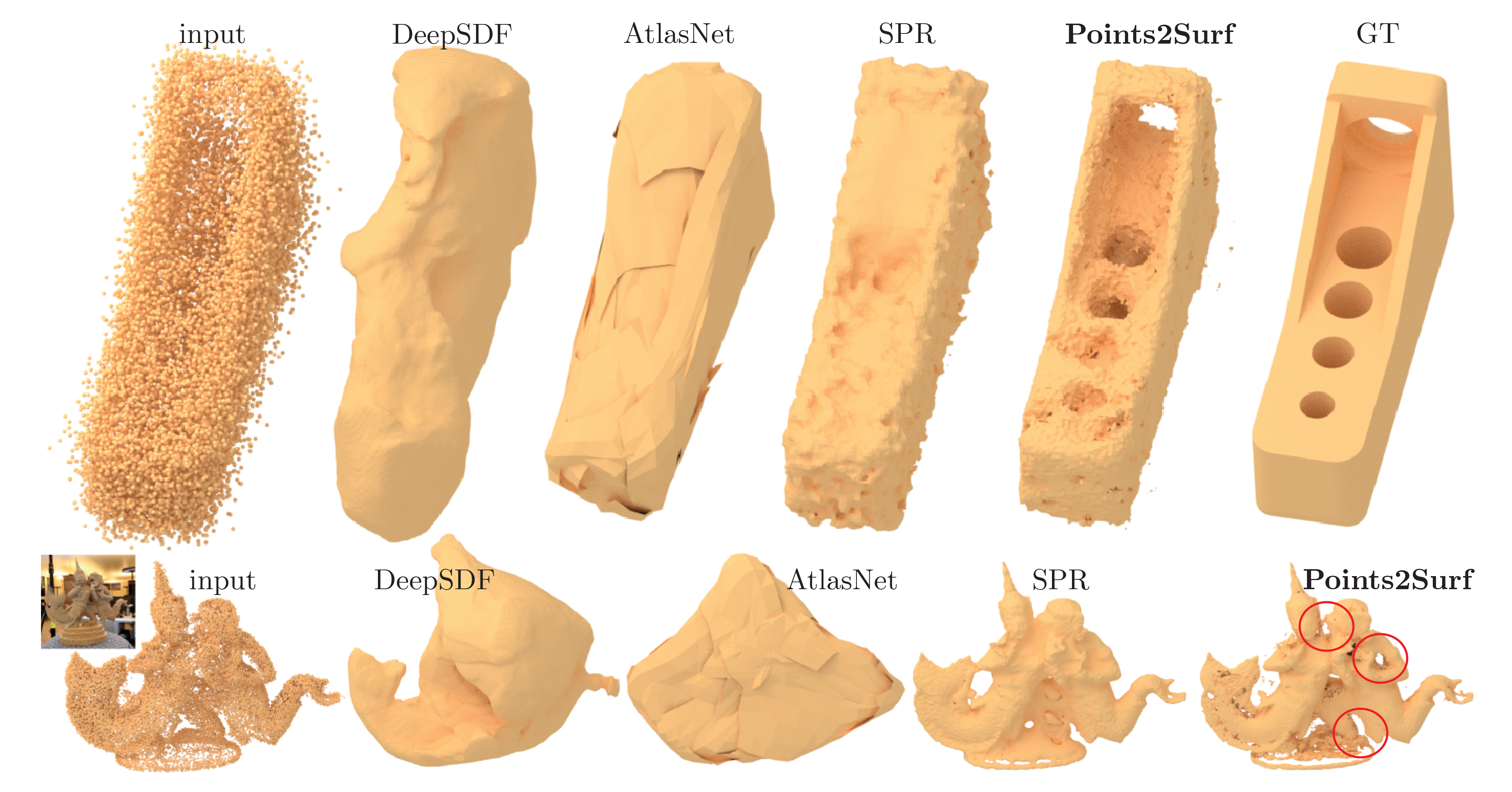
The architecture is similar to PCPNet. In contrast to other ML-based surface reconstruction methods, e.g. DeepSDF and AtlasNet, Points2Surf is patch-based and therefore independent from classes. The strongly improved generalization leads to much better results, even better than Screened Poisson Surface Reconstruction in most cases.
This code was mostly written by Philipp Erler and Paul Guerrero. This work was published at ECCV 2020.
- Python >= 3.7
- PyTorch >= 1.6
- CUDA and CuDNN if using GPU
- BlenSor 1.0.18 RC 10 for dataset generation
To get a minimal working example for training and reconstruction, follow these steps. We recommend using Anaconda to manage the Python environment. Otherwise, you can install the required packages with Pip as defined in the requirements.txt.
# clone this repo
# a minimal dataset is included (2 shapes for training, 1 for evaluation)
git clone https://github.com/ErlerPhilipp/points2surf.git
# go into the cloned dir
cd points2surf
# create a conda environment with the required packages
conda env create --file p2s.yml
# activate the new conda environment
conda activate p2s
# train and evaluate the vanilla model with default settings
python full_run.pyReconstruct meshes from a point clouds to replicate our results:
# download the test datasets
python datasets/download_datasets_abc.py
python datasets/download_datasets_famous.py
python datasets/download_datasets_thingi10k.py
python datasets/download_datasets_real_world.py
# download the pre-trained models
python models/download_models_vanilla.py
python models/download_models_ablation.py
python models/download_models_max.py
# start the evaluation for each model
# Points2Surf main model, trained for 150 epochs
bash experiments/eval_p2s_vanilla.sh
# ablation models, trained to for 50 epochs
bash experiments/eval_p2s_small_radius.sh
bash experiments/eval_p2s_medium_radius.sh
bash experiments/eval_p2s_large_radius.sh
bash experiments/eval_p2s_small_kNN.sh
bash experiments/eval_p2s_large_kNN.sh
bash experiments/eval_p2s_shared_transformer.sh
bash experiments/eval_p2s_no_qstn.sh
bash experiments/eval_p2s_uniform.sh
bash experiments/eval_p2s_vanilla_ablation.sh
# additional ablation models, trained for 50 epochs
bash experiments/eval_p2s_regression.sh
bash experiments/eval_p2s_shared_encoder.sh
# best model based on the ablation results, trained for 250 epochs
bash experiments/eval_p2s_max.shEach eval script reconstructs all test sets using the specified model. Running one evaluation takes around one day on a normal PC with e.g. a 1070 GTX and grid resolution = 256.
To train the P2S models from the paper with our training set:
# download the ABC training and validation set
python datasets/download_datasets_abc_training.py
# start the evaluation for each model
# Points2Surf main model, train for 150 epochs
bash experiments/train_p2s_vanilla.sh
# ablation models, train to for 50 epochs
bash experiments/train_p2s_small_radius.sh
bash experiments/train_p2s_medium_radius.sh
bash experiments/train_p2s_large_radius.sh
bash experiments/train_p2s_small_kNN.sh
bash experiments/train_p2s_large_kNN.sh
bash experiments/train_p2s_shared_transformer.sh
bash experiments/train_p2s_no_qstn.sh
bash experiments/train_p2s_uniform.sh
bash experiments/train_p2s_vanilla_ablation.sh
# additional ablation models, train for 50 epochs
bash experiments/train_p2s_regression.sh
bash experiments/train_p2s_shared_encoder.sh
# best model based on the ablation results, train for 250 epochs
bash experiments/train_p2s_max.shWith 4 RTX 2080Ti, we trained around 5 days to 150 epochs. Full convergence is at 200-250 epochs but the Chamfer distance doesn't change much. The topological noise might be reduced, though.
The point clouds are stored as NumPy arrays of type np.float32 with ending .npy where each line contains the 3 coordinates of a point. The point clouds need to be normalized to the (-1..+1)-range.
A dataset is given by a text file containing the file name (without extension) of one point cloud per line. The file name is given relative to the location of the text file.
To make your own dataset from meshes, place your ground-truth meshes in ./datasets/(DATASET_NAME)/00_base_meshes/. Meshes must be of a type that Trimesh can load, e.g. .ply, .stl, .obj or .off. Because we need to compute signed distances for the training set, these input meshes must represent solids, i.e be manifold and watertight. Triangulated CAD objects like in the ABC-Dataset work in most cases. Next, create the text file ./datasets/(DATASET_NAME)/settings.ini with the following settings:
[general]
only_for_evaluation = 0
grid_resolution = 256
epsilon = 5
num_scans_per_mesh_min = 5
num_scans_per_mesh_max = 30
scanner_noise_sigma_min = 0.0
scanner_noise_sigma_max = 0.05When you set only_for_evaluation = 1, the dataset preparation script skips most processing steps and produces only the text file for the test set.
For the point-cloud sampling, we used BlenSor 1.0.18 RC 10. Windows users need to fix a bug in the BlenSor code. Make sure that the blensor_bin variable in make_dataset.py points to your BlenSor binary, e.g. blensor_bin = "bin/Blensor-x64.AppImage".
You may need to change other paths or the number of worker processes and run:
python make_dataset.py
The ABC var-noise dataset with about 5k shapes takes around 8 hours using 15 worker processes on a Ryzen 7. Most computation time is required for the sampling and the GT signed distances.
If you only want to reconstruct your own point clouds, place them in ./datasets/(DATASET_NAME)/00_base_pc/. The accepted file types are the same as for meshes.
You need to change some settings like the dataset name and the number of worker processes in make_dataset_pc.py and then run it with:
python make_dataset_pc.py
In case you already have your point clouds as Numpy files, you can create a dataset manually. Put the *.npy files in the (DATASET_NAME)/04_pts/ directory. Then, you need to list the names (without extensions, one per line) in a textfile (DATASET_NAME)/testset.txt.
If you use our work, please cite our paper:
@article{ErlerEtAl:Points2Surf:ECCV:2020,
title = {{Points2Surf}: Learning Implicit Surfaces from Point Clouds},
author = {Philipp Erler and Paul Guerrero and Stefan Ohrhallinger and Michael Wimmer and Niloy J. Mitra},
year = {2020},
journal = {?},
volume = {?},
number = {?},
pages = {?},
doi = {?},
}