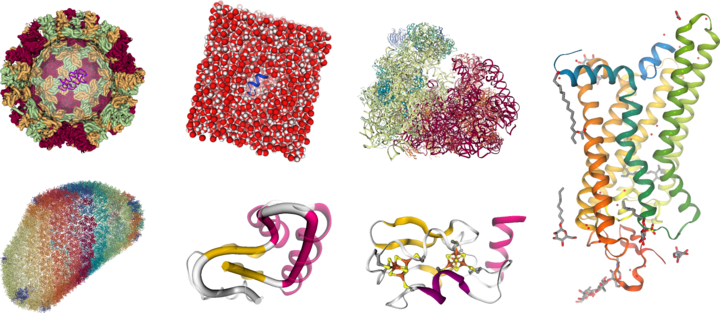
NGL Viewer is a web application for molecular visualization. WebGL is employed to display molecules like proteins and DNA/RNA with a variety of representations. Further, a server is included that enables remote access and animation of coordinate trajectories, for instance, from molecular dynamics simulations.
See it in action:
- Molecular structures (mmCIF, PDB, GRO, SDF, MOL2)
- Density volumes (MRC/MAP/CCP4)
- User interaction (mouse picking, selection language, image export)
- Coordinate trajectories (animation, server)
- Embeddable (single file, API)
This project would not be possible without recourse to many fine open-source projects. Especially the three.js project provides a great foundation. See here for a detailed list of acknowledgments.
- CODE: clearer atomnames handling for fiber creation
- FIX: residues at the end of fibers may not require all backbone atoms
- ADD: User-defined color schemes (API)
- CODE: Color handling code refactored exposing more parameters
- FIX: #7
- DOC: pull request instruction for developers
- Higher color contrast for GUI and documentation pages
- CODE: Basic support for async creation of representations (so far used for molecular surfaces and volume triangulation)
- WIP: scripting API
- CODE: chunked data loading and parsing via streamer class
- MIGRATION: Stage.loadFile signature changed
- CODE: faster autobonding of large residues (e.g. hydrated lipids)
- CODE: WebWorker support while using development and build files
- CODE: WebWorker used for decompression, parsing and surface generation
- ADD: Support for MOL2 and SDF files
The first release.
Generally MIT licensed, see the LICENSE file for details.