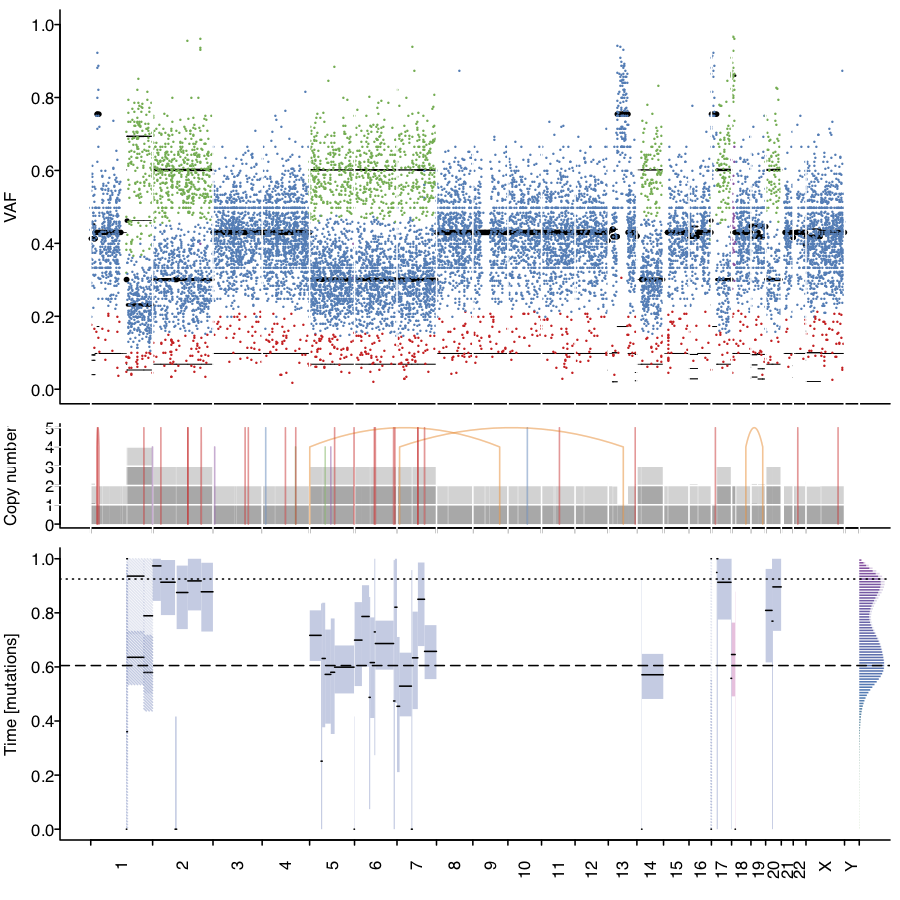
MutationTime.R is an R script to time somatic mutations relative to clonal and subclonal copy number states and calculate the relative timing of copy number gains. It has been used by the PCAWG consortium to calculate mutation times of 2,778 whole genome sequencing samples. http://dx.doi.org/10.1101/161562
MutationTime.R needs the following input objects
source("MutationTime.R")
vcf <- readVcf("myvcf.vcf") # Point mutations, needs info columns t_alt_count t_ref_count
bb <- GRanges(, major_cn= , minor_cn=, clonal_frequency=purity) # Copy number segments, needs columns major_cn, minor_cn and clonal_frequency of each segment
To run MutationTime.R
mt <- mutationTime(vcf, bb)
Mutation Annotation
head(param$D)
Classify as basic clonal states
table(classifyMutations(mt$D))
Time of copy number gains
bb$timing_param <- mt$P
bbToTime(bb)