A deep learning library focused on graph representation learning for real-world chemical tasks.
- ✅ State-of-the-art GNN architectures.
- 🐍 Extensible API: build your own GNN model and train it with ease.
- ⚗️ Rich featurization: powerful and flexible built-in molecular featurization.
- 🧠 Pretrained models: for fast and easy inference or transfer learning.
- ⮔ Read-to-use training loop based on Pytorch Lightning.
- 🔌 Have a new dataset? Goli provides a simple plug-and-play interface. Change the path, the name of the columns to predict, the atomic featurization, and you’re ready to play!
Visit https://valence-discovery.github.io/goli/.
# Install mamba if unavailable
conda install -c conda-forge mamba
# Install Goli's dependencies in a new environment named `goli_dev`
mamba env create -f env.yml -n goli_dev
# Install Goli in dev mode
conda activate goli_dev
pip install -e .mkdir ~/.venv # Create the folder for the environment
python3 -m venv ~/.venv/goli_ipu # Create the environment
source ~/.venv/goli_ipu/bin/activate # Activate the environment
# Installing the poptorch SDK. Make sure to change the path
pip install PATH_TO_SDK/poptorch-3.2.0+109946_bb50ce43ab_ubuntu_20_04-cp38-cp38-linux_x86_64.whl
# Activate poplar SDK.
source PATH_TO_SDK/enable
# Install the IPU specific and goli requirements
PACKAGE_NAME=pytorch pip install -r requirements_ipu.txt
# Install Goli in dev mode
pip install -e .To learn how to train a model, we invite you to look at the documentation, or the jupyter notebooks available here.
If you are not familiar with PyTorch or PyTorch-Lightning, we highly recommend going through their tutorial first.
See the latest changelogs at CHANGELOG.rst.
Under the Apache-2.0 license. See LICENSE.
See AUTHORS.rst.
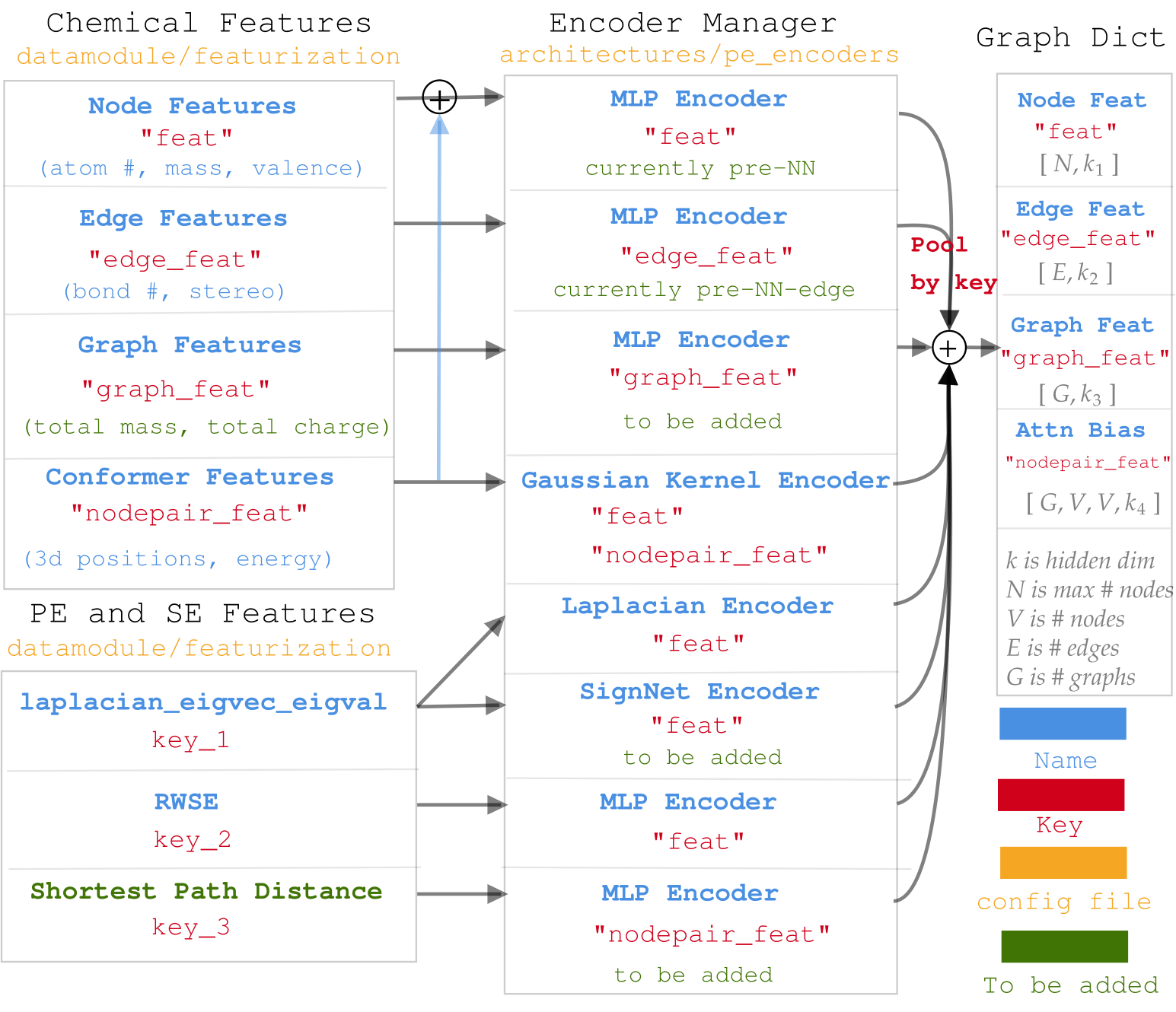
- Diagram for data processing in molGPS.
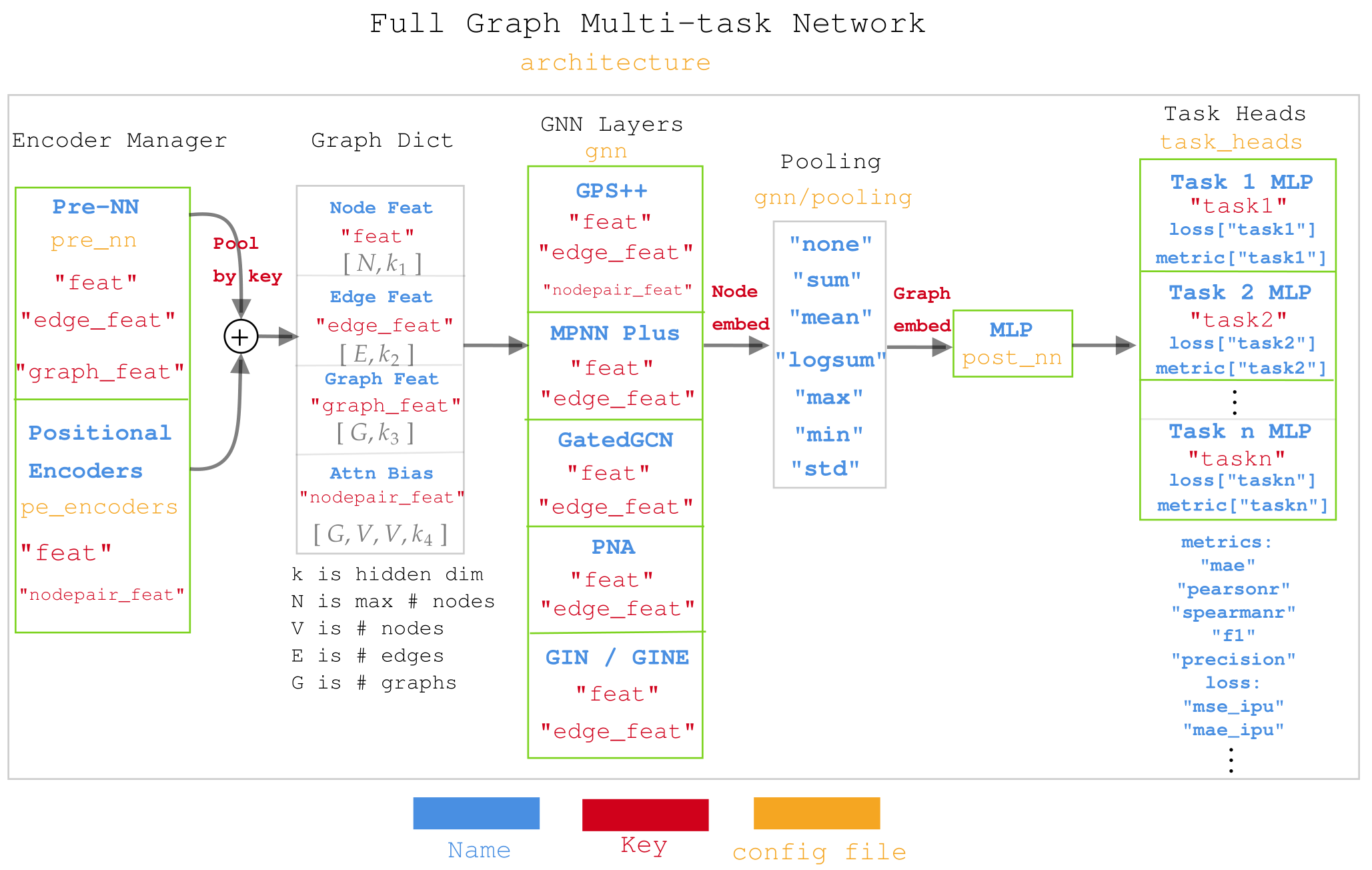
- Diagram for Muti-task network in molGPS