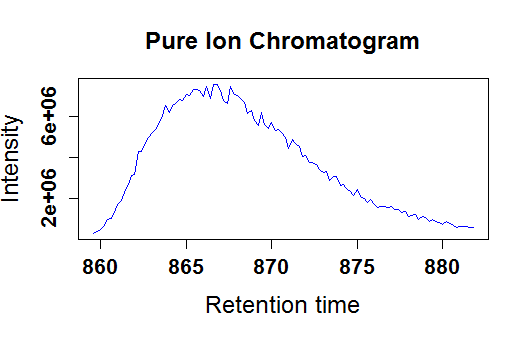
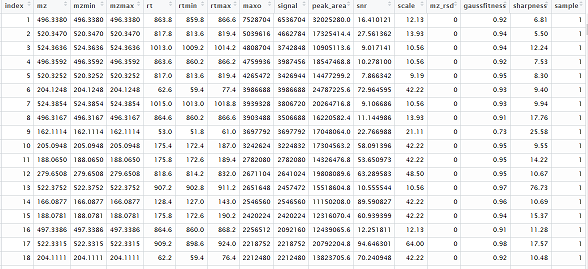
KPIC2 is an effective platform for LC-MS based metabolomics using pure ion chromatograms, which is developed for metabolomics studies. KPIC2 can detect pure ions accurately, align PICs across samples, group PICs to annotate isotope and adduct PICs, fill missing peaks and pattern recognition. High-resolution mass spectrometers like TOF and Orbitrap are more suitable.
install.packages(c("devtools","Rcpp","Ckmeans.1d.dp","foreach","doParallel","iterators","randomForest", "pracma", "caret", "ellipse", "splancs", "ggplot2", "gtools"))
source("https://bioconductor.org/biocLite.R")
biocLite(c("mzR","ropls"))
install "kopls" package from kopls_1.1.2.zip, which is embodied in KPIC2/inst
library(devtools);
install_github("hcji/KPIC2")
Extract pure ion chromatograms via optimized K-means clustering of ions in region of interest, and detect peaks of PICs. For a single sample:
library(KPIC)
filename <- 'E:/LC-MS data/example/1-1_Seg1Ev1.mzXML'
res <- getPIC(filename,roi_range=0.1,level=50000,min_snr=6,peakwidth=c(5,30))
PICplot(res,1)
For a set of samples:
files <- 'E:/LC-MS data/example'
st <- KPICset(files,roi_range=0.1,level=50000,min_snr=6,peakwidth=c(5,30))
Align PICs extracted in the last step across samples.
st <- PIAlign(st,mzSegSize=1)
Group featrues across sample based on the aligned retention time.
r.group <- PICgroup(st,tolerance=c(0.1,15),frac=0.5)
Then the combination can be used for grouping isotopic and adduct features with the main features.
r.group <- groupCombine(r.group,min_corr=0.8,type='isotope',window=10)
Summarize extracted information into a data matrix.
data.mat <- getDataMatrix(r.group,std='maxo')
After grouping, there always be peak groups that do not in-clude peaks from every sample. The cause of missing peaks mean does lies on that the peak does not exit. It may be caused by undetection or misalignment. The miss peak filling includes two steps. First, it extends the tolerance of both m/z and RT dimensions, and research from peak list of each sam-ple. Second, if there are still missing peaks, it allows feature searching from the raw data directly.
r.fillpeaks <- fillPeaks.peakfinder(data.mat,tolerance=c(0.25,25),weight=c(0.7,0.3,0.2))
r.fillpeaks <- fillPeaks.EIBPC(r.fillpeaks,tolerance=c(0.25,25),min_snr=3)
Finding the difference between two class. PLS-DA is used for the example.
x <- pretreat(r.fillpeaks)
y <- as.factor(c(rep('group1',6),rep('group2',6)))
output <- peak.analyst.plsda(x,y,ncomp=7)
For any questions, please contact: [email protected]