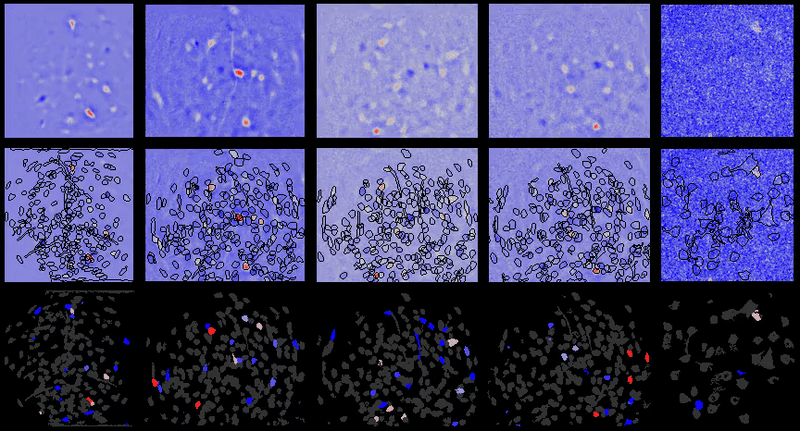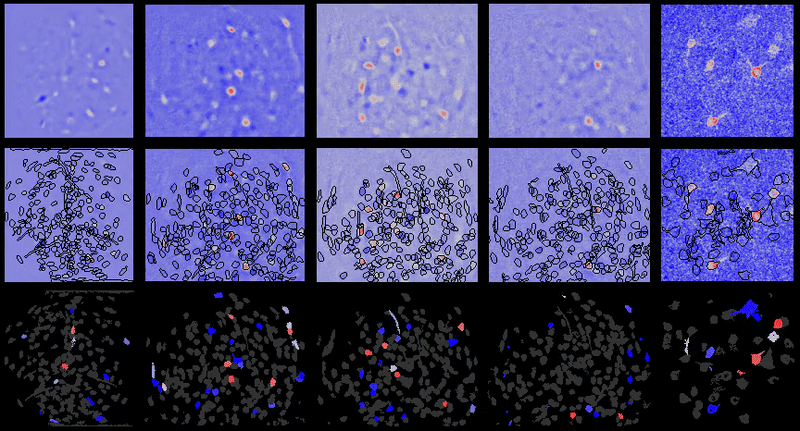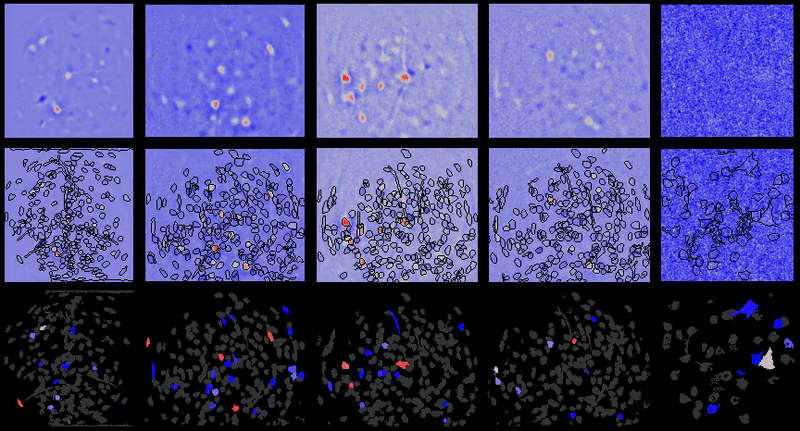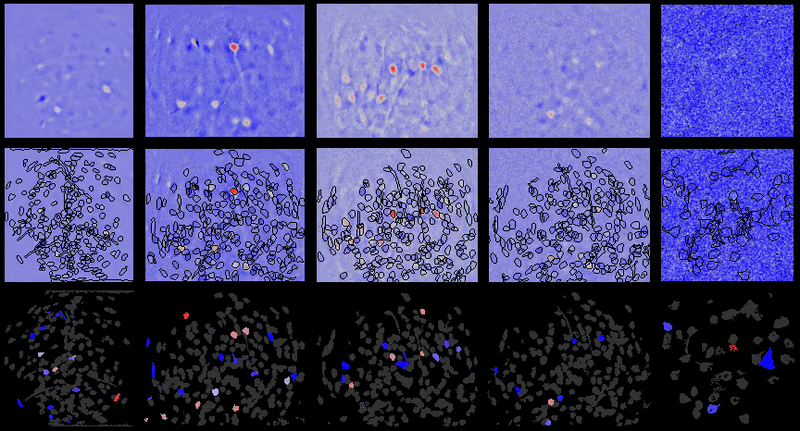
Full documentation at https://bahanonu.github.io/calciumImagingAnalysis/
CIAtah (pronounced cheetah; formerly calciumImagingAnalysis [ciapkg]) is a software package for analysis of one- and two-photon calcium imaging datasets.
Features:
- Includes a GUI with different modules to allow users to do large-scale batch analysis, accessed via the repository's
ciatahclass. - The underlying functions can be used to create GUI-less, command line-ready analysis pipelines. Functions located in
ciapkgand+ciapkgsub-folders. - Includes all major calcium imaging analysis steps: pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.), support for multiple cell-extraction methods (CELLMax, PCA-ICA, CNMF, CNMF-E, EXTRACT, etc.), manual classification via GUIs, automated cell classification (coming soon!), cross-session cell alignment, and more.
- Has several example one- and two-photon calcium imaging datasets that it will automatically download to help users test out the package.
- Includes code for determining animal position (e.g. in open-field assay).
- Supports Neurodata Without Borders data standard (see calcium imaging tutorial) for reading/writing cell-extraction (e.g. outputs of PCA-ICA, CELLMax, CNMF, CNMF-E, etc.). Supports reading and writing NWB movie files with continued integration planned.
- Requires
MATLAB.
Contact: Biafra Ahanonu, PhD (bahanonu [at] alum [dot] mit [dot] edu).
Movie processing, cell extraction, and analysis validation.
CIAtah sell sorting GUI
- Quick start guide
- Quick start (command-line)
CIAtahmain GUI notes- Acknowledgments
- References
- Questions
- License
Below are steps needed to quickly get started using the CIAtah software package in MATLAB.
- Install Clone the
CIAtahrepository (using GitHub desktop or command line) or download the repository zip and unzip (e.g. run below MATLAB command).- Point the MATLAB path to the
CIAtahroot folder (NOT@CIAtahsub-folder in the repository). - Alternatively (not recommended since lags GitHub repository), download the package from
File Exchangeusing the Add-Ons explorer in MATLAB. SeeCIAtahentry at:or https://www.mathworks.com/matlabcentral/fileexchange/75466-calciumimaginganalysis.
- Point the MATLAB path to the
% Optional: this will set MATLAB working folder to the default user path. Make sure you have read/write permissions.
cd(userpath);
% Download and unzip current repository
unzip('https://github.com/bahanonu/calciumImagingAnalysis/archive/master.zip');
% Make CIAtah the working folder
cd('calciumImagingAnalysis-master')- Run
CIAtahusing the below MATLAB commands. Callobj;each time you want to go back to the main GUI.
% Loads the class into an object for use in this session
obj = ciatah;
% Runs routines to check dependencies and help user get setup.
obj.setup;
% Open the class menu (always type `obj` then enter load the class/modules menu)
obj % then hit enter, no semicolon!- Afterwards, likely want to run
modelAddNewFoldersmodule first in order to add folders containing imaging data to the current class object. - [Optional] Users on Windows systems should download
Everything(https://www.voidtools.com/). It is a very useful and extremely fast search engine for files and folders on a computer that can allow users to quickly get full paths for lists of folders that need to be analyzed inCIAtah. - [Optional] Users who want to analyze data via the command line can run
edit ciapkg.demo.cmdLinePipelineand run each segment of code there to see what commands are needed to perform each step. It assumes you have already runexample_downloadTestData.
After downloading CIAtah and running the setup as above, users interested in command-line processing can open up the example M-file by running the below command. By running individual code-block cells, users are guided from pre-processing through cell-extraction to cross-session analysis.
edit ciapkg.demo.cmdLinePipeline- Run
obj;in the command window to see the main GUI. - All main decisions for choosing a module to run, deciding on a cell-extraction algorithm, and which folders to analyze are in a single window.
- The GUI will real-time update the selected folders based on the selections in the subject, assay, and folder filter areas.
- Sections not relevant for a specific module are grayed out.
- Tab to cycle through selection areas. Green background is the currently selected area, dark gray background is area that had previously been selected but is not the active area, and white background is for areas that have not been selected yet.
- Hover mouse over module names for tooltip that gives additional information.
For example, selecting middle two assays automatically changes selection in Loaded folders section.
Certain sections become available when user selects the appropriate module (e.g. cell-extraction module available when selecting modelExtractSignalsFromMovie).
- See additional details on the Processing calcium imaging data page for running the full processing pipeline.
- Settings used to pre-process imaging movies (
modelPreprocessMoviemodule) are stored inside the processed HDF5 movie to allowCIAtahto load them again later. - To force load all directories, including most external software packages (in
_external_programsfolder), typeciapkg.loadAllDirs;into MATLAB command line. This is most relevant when you need to access specific functions in an outside repository that are normally hidden until needed. - When issues are encountered, first check the Common issues and fixes page to see if a solution is there. Else, submit a new issue or email Biafra (bahanonu [at] alum [dot] mit [dot] edu).
- There are two sets of test data that are downloaded:
- Single session analysis:
data\2014_04_01_p203_m19_check01_rawcan be used to test the pipeline until the cross-session alignment step. - Batch analysis:
data\batchcontains three imaging sessions that should be processed and can then be used for the cross-session alignment step. Users should try these sessions to get used to batched analysis.
- Single session analysis:
- For Fiji dependency, when path to
Miji.m(e.g.\Fiji.app\scriptsfolder) is requested, likely incalciumImagingAnalysis\_external_programs\FIJI_FOLDER\Fiji.app\scriptswhereFIJI_FOLDERvaries depending on OS, unless the user requested a custom path or on OSX (in which case, find Fiji the install directory).- If you run into Java heap space memory errors when Miji tries to load Fiji in MATLAB, make sure "java.opts" file is in MATLAB start-up folder or that the
CIAtahroot folder is the MATLAB start-up folder (instructions on changing).
- If you run into Java heap space memory errors when Miji tries to load Fiji in MATLAB, make sure "java.opts" file is in MATLAB start-up folder or that the
CIAtahoften uses regular expressions to find relevant movie and other files in folders to analyze.- For example, by default it looks for any movie files in a folder containing
concat, e.g.concat_recording_20140401_180333.h5(test data). If you have a file calledrawData_2019_01_01_myInterestingExperiment.aviand all your raw data files start withrawData_then change the regular expression torawData_when requested by the repository. SeesetMovieInfomodule to change after adding new folders.
- For example, by default it looks for any movie files in a folder containing
CIAtahgenerally assumes users have imaging data associated with one imaging session and animal in a given folder. Follow folder naming conventions in Data formats for the best experience.- External software packages are downloaded into
_external_programsfolder and should be placed there if done manually.
Users can alternatively run setup as below.
% Run these commands in MATLAB to get started.
% Loads all directories
loadBatchFxns;
% Loads the class into an object for use in this session
obj = calciumImagingAnalysis;
% Download and load dependent software packages into "_external_programs" folder.
% Also download test data into "data" folder.
% Normally only need to one once after first downloading calciumImagingAnalysis package.
obj.loadDependencies;
% Add folders containing imaging data.
obj.modelAddNewFolders;
% [optional] Set the names calciumImagingAnalysis will look for in each folder
obj.setMovieInfo;
% Open class menu to pick module to run.
obj.runPipeline; % then hit enter!Thanks to Jones G. Parker, PhD (https://parker-laboratory.com/) for providing extensive user feedback during development of the calciumImagingAnalysis software package.
Additional thanks to Drs. Jesse Marshall, Jérôme Lecoq, Tony H. Kim, Hakan Inan, Lacey Kitch, Maggie Larkin, Elizabeth Otto Hamel, Laurie Burns, and Claudia Schmuckermair for providing feedback, specific functions, or helping develop aspects of the code used in the calciumImagingAnalysis software package.
Please cite Corder*, Ahanonu*, et al. 2019 Science publication or the Ahanonu, 2018 Zenodo release if you used the software package or code from this repository to advance/help your research:
@article{corderahanonu2019amygdalar,
title={An amygdalar neural ensemble that encodes the unpleasantness of pain},
author={Corder, Gregory and Ahanonu, Biafra and Grewe, Benjamin F and Wang, Dong and Schnitzer, Mark J and Scherrer, Gr{\'e}gory},
journal={Science},
volume={363},
number={6424},
pages={276--281},
year={2019},
publisher={American Association for the Advancement of Science}
}@misc{biafra_ahanonu_2018_2222295,
author = {Biafra Ahanonu},
title = {{calciumImagingAnalysis: a software package for
analyzing one- and two-photon calcium imaging
datasets.}},
month = December,
year = 2018,
doi = {10.5281/zenodo.2222295},
url = {https://doi.org/10.5281/zenodo.2222295}
}Please email any additional questions not covered in the repository to bahanonu [at] alum [dot] mit [dot] edu or open an issue.
Copyright (C) 2013-2020 Biafra Ahanonu
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.