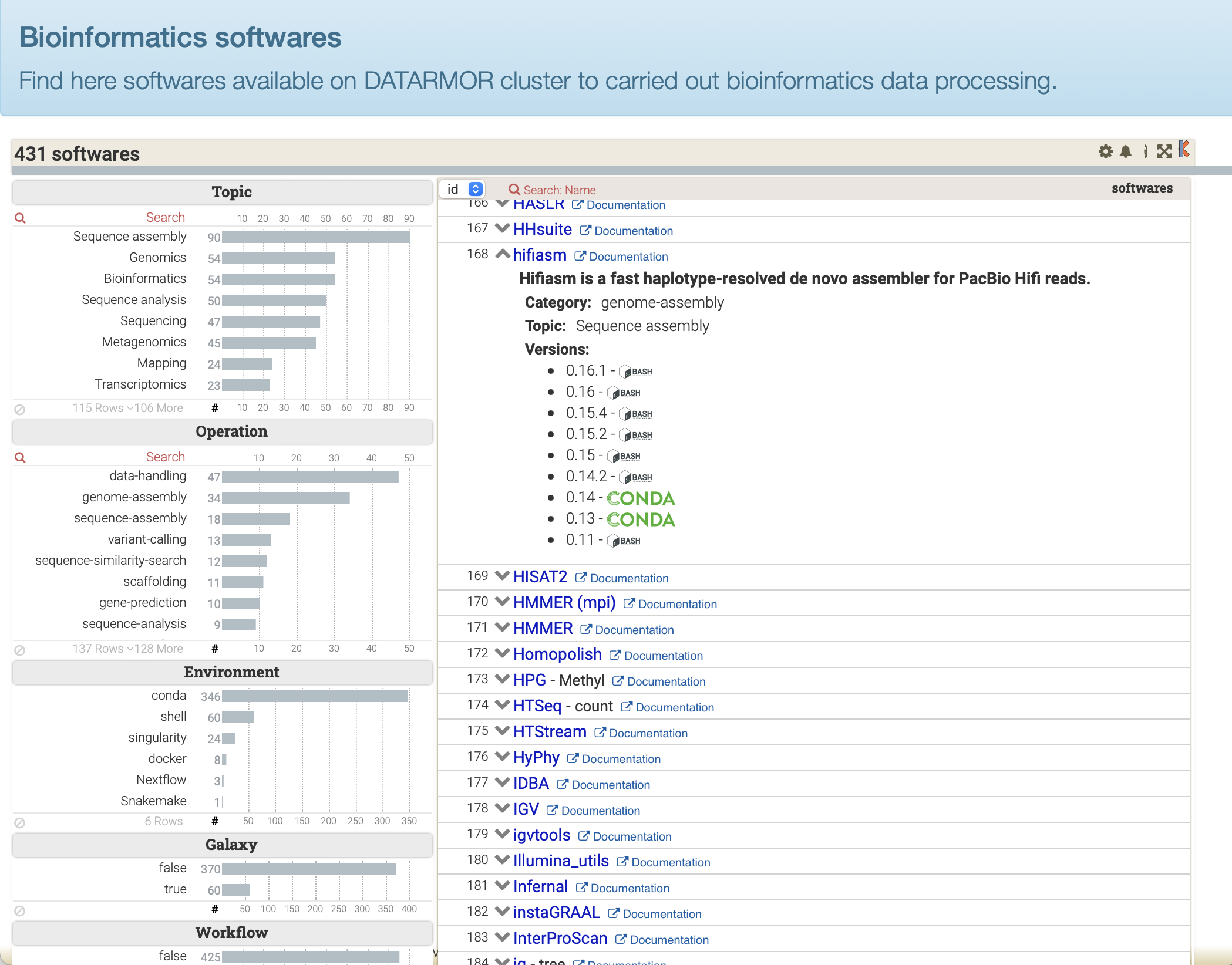
ToolDirectory provides a convenient tool to display list of softwares in a webpage along with dynamic data filtering capabilities.
You can test our public demo
The only step you have to achieve consists in creating a specific CSV file, listing all your softwares along with some metadata, such as EDAM terms, links to documentation, etc. An example is provided in test file Softwares.csv.
Then, review this fully running example of ToolDirectory Viewer tool : ToolDirectory Viewer at work, you'll see how a Softwares.csv file is used.
That's all folks to use the Viewer.
Now, to create the CSV file requested by the Viewer in a fully automated way, ToolDirectory provides a few tools, hereafter presented.
ToolDirectory provides tools to handle a catalogue of softwares relying on a particular way of installating softwares on your system (see below, Expected directory structure).
ToolDirectory is a Python 3.x program. It also requires the following package:
- Requests v2.25.1
conda create -p tooldir -c anaconda requests=2.25.1
Web rendering relies on the open-source version of Keshif data visualisation. We provided Katalog, a lightweight version specifically designed for ToolDirectory and DataDirectory.
$ tooldir -h
usage: tooldir <command> [<args>]
The available commands are:
create Create tool properties
status Set installation status of tool/version
kcsv Create csv for visualisationToolDirectory expects a directory structure with the following constraints:
- <install-dir>/<tool>/<version>/
Or, with modules architecture:
- <install-dir>/<tool>/
Here is an example:
/path/to/softwares
├── blast
│ ├── 2.2.31 #folder or module
│ └── 2.6.0
├── plast
│ └── 2.3.2
├── beedeem
│ └── 4.3.0
.../...
tooldir create -n <toolname> -v <version> -o <username>You will need Katalog, a lightweight version of Keshif specifically designed for ToolDirectory and DataDirectory.
git clone https://gitlab.ifremer.fr/bioinfo/katalog.git /foo/bar/www/tooldirectoryThen, generate the software list:
tooldir kcsv -p /path/to/softwares/ -o /foo/bar/www/tooldirectory/Softwares.csvYou can use a crontab to automatically update the software listing.
Tool Directory is released under the terms of the Apache 2 license.