Source United States Department of Health and Human Services Health Resources and Services Administration
HRSA Data Warehouse: https://datawarehouse.hrsa.gov/data/datadownload.aspx
Summary
The Area Health Resource File (ARF) is a health resource information database containing more than 6,000 variables for each of the nation's counties. ARF contains information on health facilities, health professions, measures of resource scarcity, health status, economic activity, health training programs, and socioeconomic and environmental characteristics.
Detailed information about the AHRF dataset
See the files in the metadata subdirectory:
- AHRF User Guide: http://bit.ly/2ngOWBw
- AHRF Technical Documentation (Data Dictionary): http://bit.ly/2mY3pzS
- Health Care Disparities in Rural Areas Fact Sheet: http://bit.ly/2nrsPZw
- National Health Care Quality Report: http://bit.ly/2nMRTvf
This repository is designed to download and install the datasets as an R package. The size of ahrf_county.rda is 24.2 MB, so it might take a while to install and load into memory.
# install.packages("devtools")
devtools::install_github("olyerickson/ahrf2016r")
# To uninstall the package, use:
# remove.packages("ahrf2016r")
The data directory contains the full county data, plus training (75% of observations) and test (25%) subsets:
ahrf_county.rda
ahrf_county_train.rda
ahrf_county_test.rda
NOTE:
- Only
ahrf_county_train.rdawill be available before and during the 2017 RPI Datathon (25-26 Mar 2017) ahrf_county.rdaandahrf_count_test.rdawill become available after 26 Mar 2017
There are 3230 rows and 6921 columns in the full county file (wide format):
NOTE: Substitute ahrf_county_train as needed...
library(dplyr, warn.conflicts = FALSE)
dim(ahrf2016r::ahrf_county)
#> [1] 3230 6921
What columns mention "beds"?
> beds_rows <- ahrf_county_layout[grep("beds", ahrf_county_layout$label_2, ignore.case=T),]
> View(beds_rows)
What columns mention "population"?
> pop_rows <- ahrf_county_layout[grep("population", ahrf_county_layout$label_2, ignore.case=T),]
> View(pop_rows)
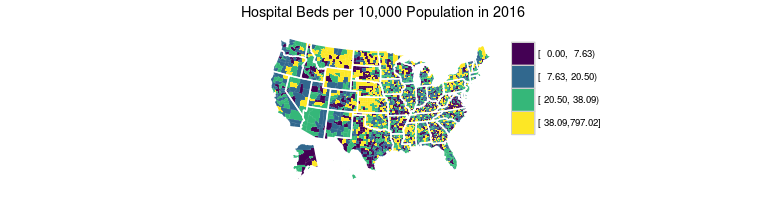
County-level hospital beds in 2016
> ahrf2016r::ahrf_county %>%
select(county = F04437,
fips = F00002,
beds_2016 = F0892113,
pop_2016 = F1198415) %>%
mutate(beds_2016 = as.integer(beds_2016),
pop_2016 = as.integer(pop_2016),
beds_2016_p10k = beds_2016 / pop_2016 * 10000) -> beds
> beds
# A tibble: 3,230 × 5
county fips beds_2016 pop_2016 beds_2016_p10k
<chr> <chr> <int> <int> <dbl>
1 Autauga, AL 01001 50 55347 9.033913
2 Baldwin, AL 01003 364 203709 17.868626
3 Barbour, AL 01005 47 26489 17.743214
4 Bibb, AL 01007 20 22583 8.856219
5 Blount, AL 01009 40 57673 6.935654
6 Bullock, AL 01011 54 10696 50.486163
7 Butler, AL 01013 83 20154 41.182892
8 Calhoun, AL 01015 458 115620 39.612524
9 Chambers, AL 01017 188 34123 55.094804
10 Cherokee, AL 01019 45 25859 17.402065
# ... with 3,220 more rows
> summary(beds$beds_2016)
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.0 17.0 50.0 294.6 181.5 25310.0 7
> summary(beds$pop_2016)
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
89 11280 25960 100900 66750 10170000 9
> summary(beds$beds_2016_p10k)
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
0.000 7.367 20.090 30.680 37.250 797.000 14
> quantile(beds$beds_2016_p10k, na.rm = TRUE)
0% 25% 50% 75% 100%
0.000000 7.366996 20.093600 37.248540 797.016672
Geographic Distribution of Hospital Beds
beds$beds_2016_dist = Hmisc::cut2(beds$beds_2016_p10k, cuts = c(7.63, 20.50, 38.09))
# devtools::install_github("jjchern/usmapdata")
usmapdata::county %>%
left_join(beds, by = c("id" = "fips")) %>%
mutate(region = id) -> beds_map
usmapdata::state %>%
mutate(region = id) -> state_map
library(ggplot2)
ggplot() +
geom_map(data = beds_map, map = beds_map,
aes(x = long, y = lat, map_id = id, fill = beds_2016_dist),
colour = alpha("white", 0.1), size=0.2) +
geom_map(data = state_map, map = state_map,
aes(x = long, y = lat, map_id = region),
colour = "white", fill = "NA") +
coord_map("albers", lat0 = 30, lat1 = 40) +
viridis::scale_fill_viridis(discrete=TRUE, option = "D") +
ggtitle("Hospital Beds per 10,000 Population in 2016") +
ggthemes::theme_map() +
theme(legend.position = c(.85, .3),
legend.title=element_blank())