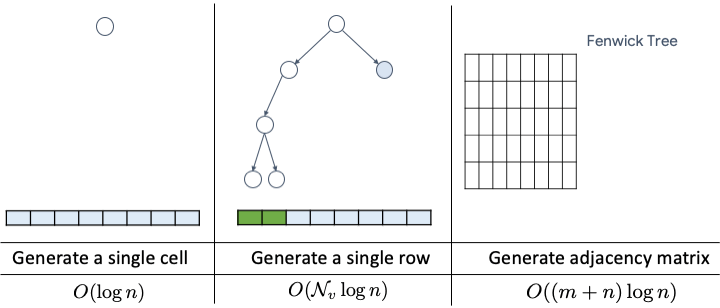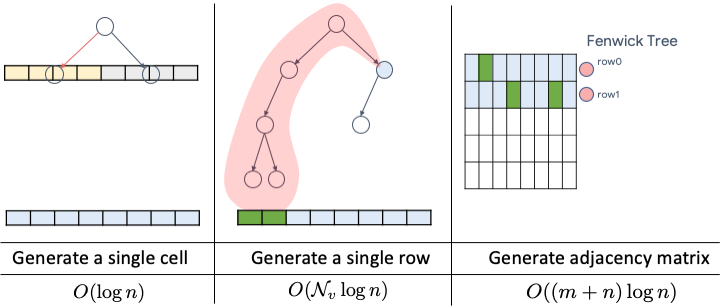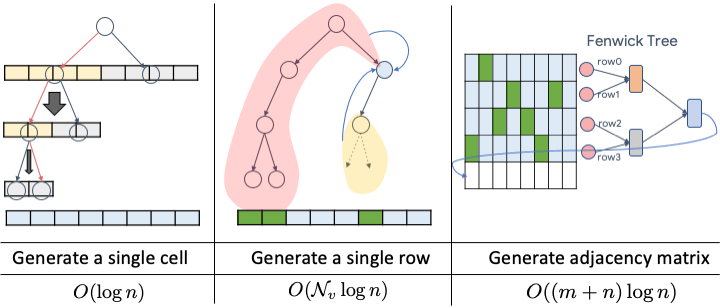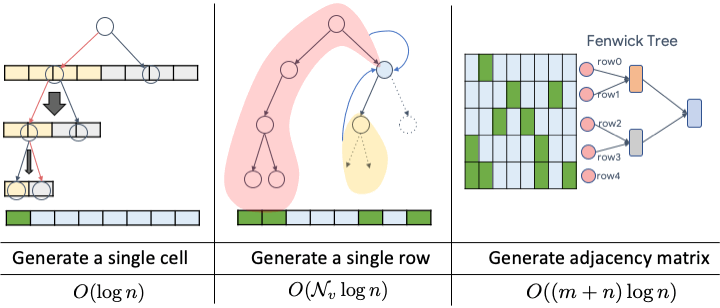
Paper available at: https://arxiv.org/pdf/2006.15502.pdf
Upon using this codebase, please cite the paper:
@article{dai2020scalable,
title={Scalable Deep Generative Modeling for Sparse Graphs},
author={Dai, Hanjun and Nazi, Azade and Li, Yujia and Dai, Bo and Schuurmans, Dale},
journal={arXiv preprint arXiv:2006.15502},
year={2020}
}
New feature enabled: now BiGG is able to generate graphs with node/edge features, fully in an autoregressive way.
Please refer to bigg/extension/customized_models.py for an illustration of how to customize your own model with graph node/edge features.
One can directly run the script bigg/extension/run_featured.sh once the graph data (list of networkx graphs stored in a pickle file) is provided.
Navigate to the root of project, and perform:
pip install -e .
The installation requires both gcc and CUDA toolkit (if gpu is enabled)
If you want to reproduce the benchmark results, please also install GRAN and G2SAT and add them into your PYTHONPATH.
Please organize this project according to following structure:
bigg/
|___bigg/ # source code
| |___common # common implementations
| |___...
|
|___setup.py
|
|___data/ # raw data / cooked data
| |___DD/
| | |__DD_A.txt
| | |__...
| |___G2SAT
| | |__lcg_stats.csv
| | |__train_formulas/
| | |__test_formulas/
| |___...
|
|___results # pretrained model and generated graphs
| |___DD/
| |___...
|
|...
Besides the synthetic data, we use the publicly avaiable benchmark datasets.
-
Data from GRAN
Please download the raw data from https://github.com/lrjconan/GRAN/tree/master/data and put folders into data/.
-
Data from G2SAT
Please download the raw data from https://github.com/JiaxuanYou/G2SAT/tree/master/dataset and put them under data/.
Make sure the folder structure is consistent.
You can find the pretrained models via https://console.cloud.google.com/storage/browser/research_public_share/bigg_icml2020
To use these models, please download and put the folders under results/.
First let's cook the data using following steps
cd bigg/data_process
./run_syn_datagen.sh
The above script will generate the train/val/test split of the corresponding dataset. Please modify the g_type field for different datasets, and ordering for different node orderings.
Make sure the model dumps are downloaded and in the specific location.
cd bigg/experiments/synthetic/scripts
./run_lobster.sh -phase test -epoch_load -1
The above script will generate graphs in a pickle file in the same folder as the corresponding model dump. You can also try the scripts for other types of graphs.
To train from scratch, simply run the script directly
./run_grid.sh
We use step-decay for the learning rate. When the training loss gets plateau, you need to reduce the learning rate:
./run_grid.sh -epoch_load EPOCH -learning_rate 1e-5
You can specify the EPOCH number to load to continue training.
First follow the instruction in https://github.com/JiaxuanYou/G2SAT to process the formulas into edge_list. You are supposed to get the set of edge list under the data folder:
data/
|___G2SAT
|__train_formulas/
|__train_set/
|__test_formulas/
|__...
After that, please cook the data using the following script
cd bigg/data_process
./run_sat_gen.sh
You can also modify the folder_name to cook different data portion.
Make sure the model dumps are downloaded and in the specific location.
cd bigg/experiments/sat_graphs
./run_full.sh -eval_folder train -epoch_load -1 -greedy_frac 0.9
The above script evaluates the consistency with training set, with a mixed sampling distribution between greedy and multinomial one.
After generation, you are supposed to get the formula folder under results/sat-train/blk-500-b-1/train-pred_formulas-e--1-g-0.00/.
Please follow the instructions in G2SAT to evaluate the statistics of your generated formulas.
Simply run the run_full.sh or run_sub.sh script directly, depends on the training set.
Note that the graphs are large, so please adjust the blksize in the scripts to better fit into GPU memory.
We also demonstrate the model parallelism for BiGG. You don't have to use this option and all the experiments can be reproduced in a single GPU, but this can be 2x faster if you have multiple GPUs.
cd bigg/experiments/synthetic
./demo_model_parallelism.sh
You can tune the block size (blksize), number of processes (should be <= num of available gpus)