Powerful statistics for VCF files
vcfstats also requires R with ggplot2 to be installed.
If you are doing pie chart, ggrepel is also required.
pip install vcfstatsvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1) ~ CONTIG' \
--title 'Number of variants on each chromosome' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1) ~ CONTIG' \
--title 'Number of variants on each chromosome (modified)' \
--config examples/config.toml \
--ggs 'scale_x_discrete(name ="Chromosome", \
limits=c("1","2","3","4","5","6","7","8","9","10","X")) + \
ylab("# Variants")'vcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1) ~ CONTIG[1,2,3,4,5]' \
--title 'Number of variants on each chromosome (first 5)' \
--config examples/config.toml
# or
vcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1) ~ CONTIG[1-5]' \
--title 'Number of variants on each chromosome (first 5)' \
--config examples/config.toml
# or
# require vcf file to be tabix-indexed.
vcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1) ~ CONTIG' \
--title 'Number of variants on each chromosome (first 5)' \
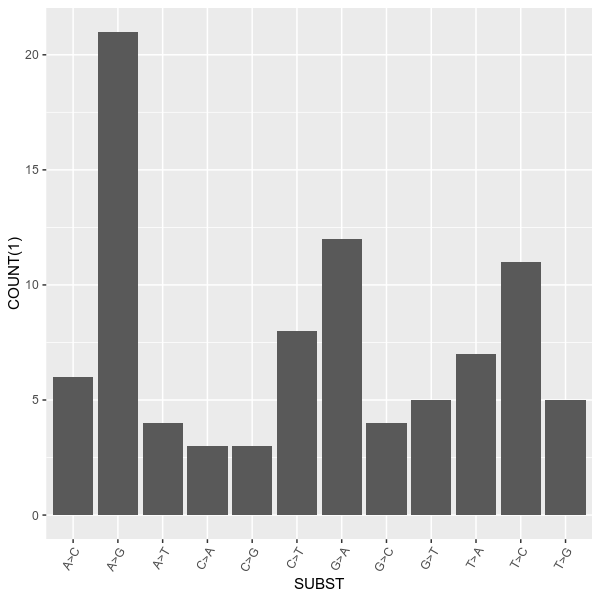
--config examples/config.toml -r 1 2 3 4 5vcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1, VARTYPE[snp]) ~ SUBST[A>T,A>G,A>C,T>A,T>G,T>C,G>A,G>T,G>C,C>A,C>T,C>G]' \
--title 'Number of substitutions of SNPs' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1, VARTYPE[snp]) ~ SUBST[A>T,A>G,A>C,T>A,T>G,T>C,G>A,G>T,G>C,C>A,C>T,C>G]' \
--title 'Number of substitutions of SNPs (passed)' \
--config examples/config.toml \
--passed# using a dark theme
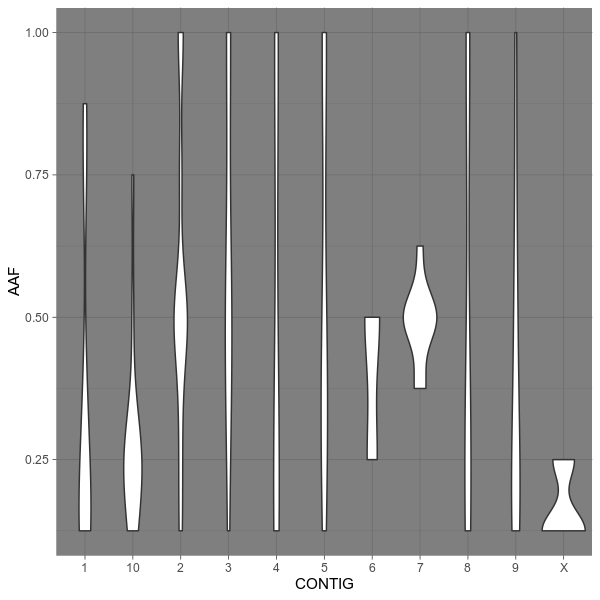
vcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'AAF ~ CONTIG' \
--title 'Allele frequency on each chromosome' \
--config examples/config.toml --ggs 'theme_dark()'vcfstats --vcf examples/sample.vcf \
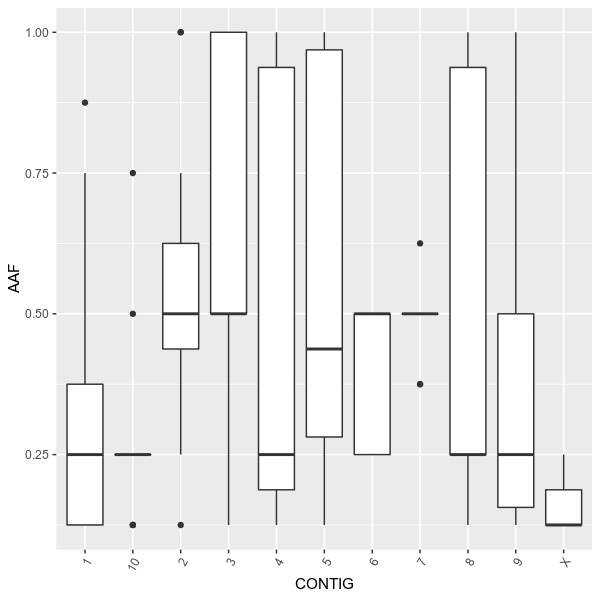
--outdir examples/ \
--formula 'AAF ~ CONTIG' \
--title 'Allele frequency on each chromosome (boxplot)' \
--config examples/config.toml \
--figtype boxplotYou can plot the distribution, using density plot or histogram
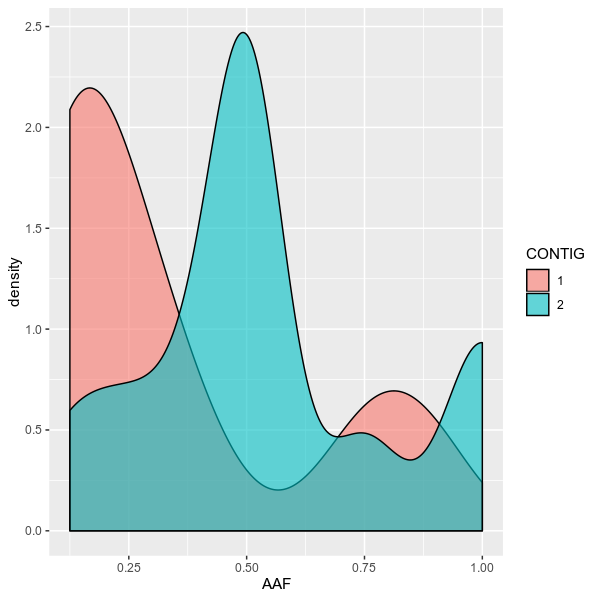
vcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'AAF ~ CONTIG[1,2]' \
--title 'Allele frequency on chromosome 1,2' \
--config examples/config.toml \
--figtype densityvcfstats --vcf examples/sample.vcf \
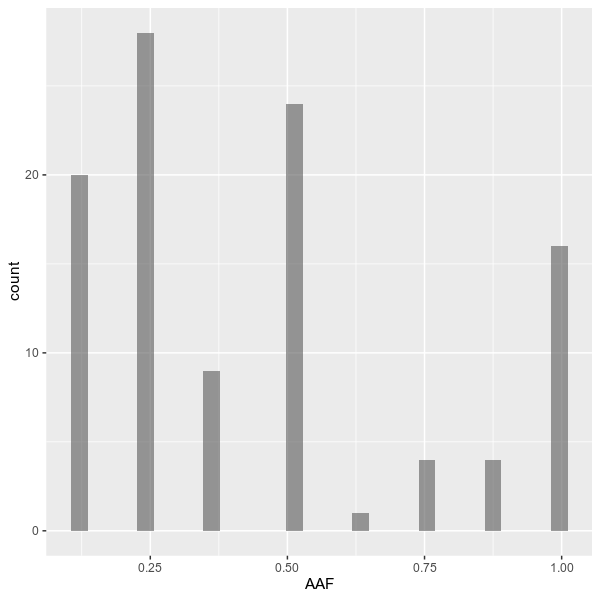
--outdir examples/ \
--formula 'AAF ~ 1' \
--title 'Overall allele frequency distribution' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'AAF[0.05, 0.95] ~ 1' \
--title 'Overall allele frequency distribution (0.05-0.95)' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1, group=VARTYPE) ~ CHROM' \
--title 'Types of variants on each chromosome' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1, group=VARTYPE) ~ CHROM[1]' \
--title 'Types of variants on each chromosome 1' \
--config examples/config.toml \
--figtype pievcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1, group=VARTYPE) ~ 1' \
--title 'Types of variants on whole genome' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'COUNT(1, group=GTTYPEs[HET,HOM_ALT]{0}) ~ CHROM' \
--title 'Mutant genotypes on each chromosome (sample 1)' \
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'MEAN(GQs{0}) ~ MEAN(DEPTHs{0}, group=CHROM)' \
--title 'GQ vs depth (sample 1)' \
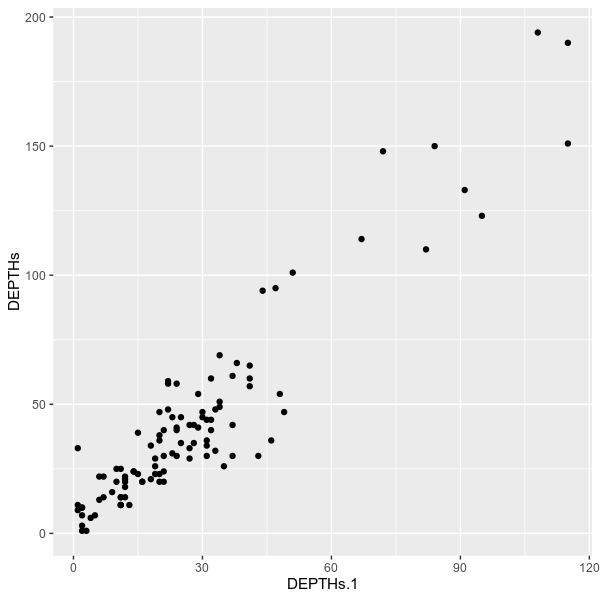
--config examples/config.tomlvcfstats --vcf examples/sample.vcf \
--outdir examples/ \
--formula 'DEPTH{0} ~ DEPTH{1}' \
--title 'Depths between sample 1 and 2' \
--config examples/config.toml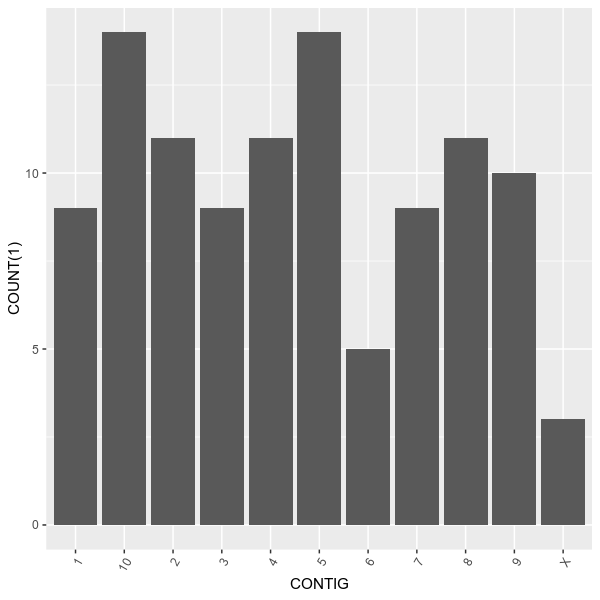
.col.png)
.col.png)
.col.png)
.histogram.png)



.col.png)
.scatter.png)