Protein structures prediction is important because the accuracy of protein structures influence how our understanding of its function and its interactions with other molecules, which can help to design new drugs to target specific molecular interactions.
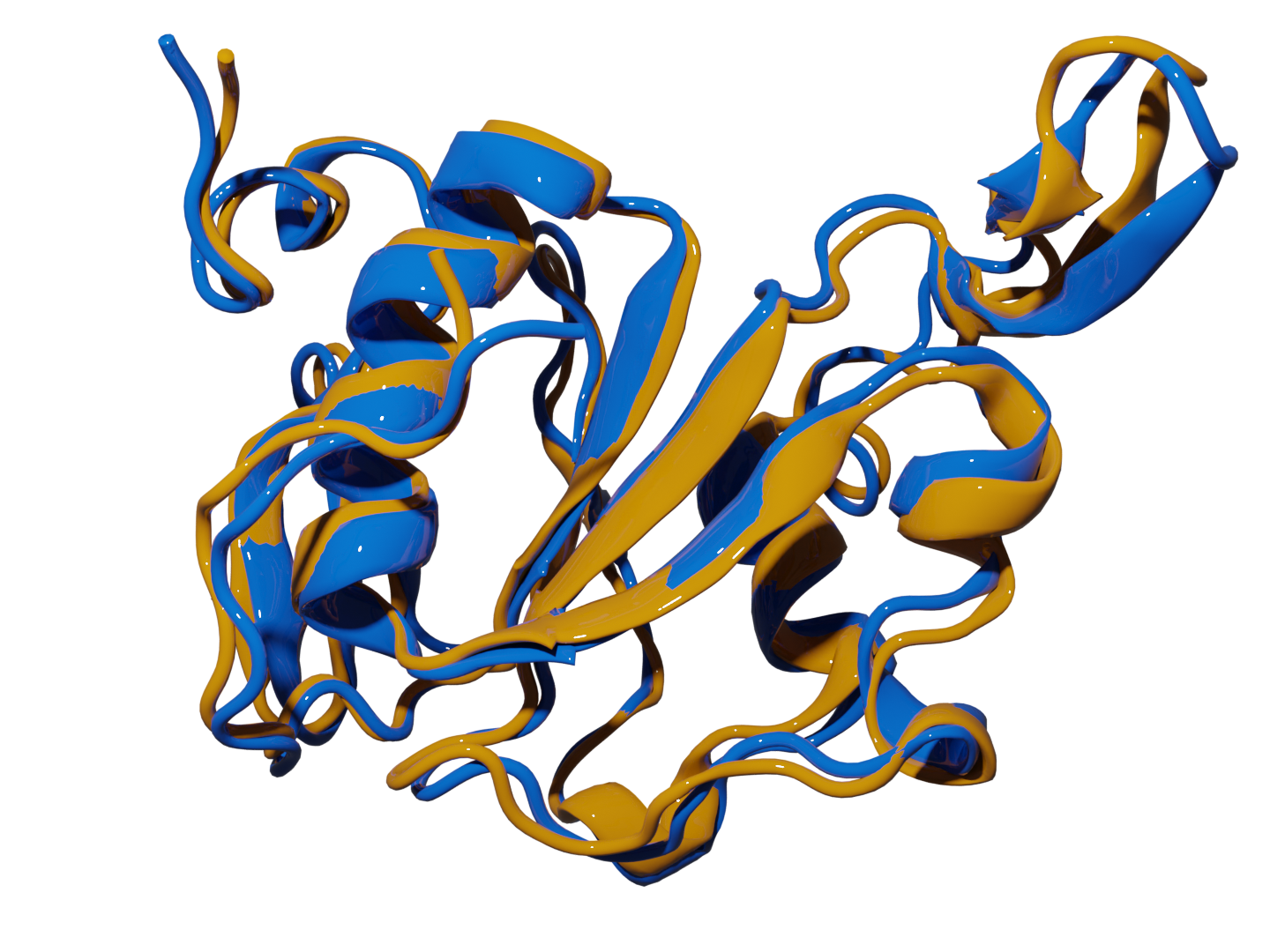
Fig 1. Superposition of the most representative structures found with extended indices (yellow) and experimental native structures (blue) of 2k2e.Protein Retrieval via Integrative Molecular Ensembles (PRIME) is a novel algorithm that predicts the native structure of a protein from simulation or clustering data. This repo contains six different ways of determining the native structure of biomolecules from simulation or clustering data. These methods perfectly mapped all the structural motifs in the studied systems and required unprecedented linear scaling.
   
git clone https://github.com/lexin-chen/PRIME.git
cd PRIME
requires Python 3.6+ and the following packages: MDAnalysis, numpy, and matplotlib.
modules contains the functions required to run the algorithm. sample_clusters/clusttraj.c* contains sample clustering files prepared through CPPTRAJ Hierarchical clustering. new_clusters/normalize.py normalizes clustering files through min-max normalization. similarity.py generates a similarity dictionary from running the protein refinement method.
The following tutorial will guide you through the process of determining the native structure of a biomolecule using the PRIME algorithm. If you already have clustered data, you can skip to Step 3.
Preparation for Molecular Dynamics Trajectory
Prepare a valid topology file (e.g. .pdb, .prmtop), trajectory file (e.g. .dcd, .nc), and the atom selection. This step will convert a Molecular Dynamics trajectory to a numpy ndarray. Make sure the trajectory is already aligned and/or centered if needed!
Step-by-step tutorial can be found in the scripts/inputs/preprocessing.ipynb.
In this example, we will use k-means clustering to assign labels to the clusters and the number of clusters will be 20. Any clustering method can be used as long as the data is clustered (e.g. DBSCAN, Hierarchical Clustering). Please check out MDANCE for more clustering methods!
scripts/nani/assign_labels.py will assign labels to the clusters using k-means clustering
# System info - EDIT THESE
input_traj_numpy = '../../example/aligned_tau.npy'
N_atoms = 50
sieve = 1
# K-means params - EDIT THESE
n_clusters = 20
output_dir = 'outputs'
input_traj_numpy is the numpy array prepared from step 1.
N_atoms is the number of atoms used in the clustering, should be same as atom selection in step 1.
sieve takes every sieveth frame from the trajectory for analysis.
n_clusters is the number of clusters for labeling.
output_dir is the directory where the output files will be saved.
python assign_labels.py- csv file containing the cluster labels for each frame.
- csv file containing the population of each cluster.
With already clustered data, scripts/normalization/normalize.py Normalize the trajectory data between
# System info - EDIT THESE
input_top = '../../example/aligned_tau.pdb'
unnormed_cluster_dir = '../clusters/outputs/clusttraj_*'
output_base_name = 'normed_clusttraj'
atomSelection = 'resid 3 to 12 and name N CA C O H'
n_clusters = 10
input_top is the topology file used in the clustering.
unnormed_cluster_dir is the directory where the clustering files are located from step 2.
output_base_name is the base name for the output files.
atomSelection is the atom selection used in the clustering.
n_clusters is the number of clusters used in the PRIME. If number less than total number of cluster, it will take top n number of clusters.
python normalize.pynormed_clusttraj.c*.npyfiles, normalized clustering files.normed_data.npy, appended all normed files together.
scripts/prime/exec_similarity.py generates a similarity dictionary from running PRIME.
-h- for help with the argument options.-m- methods, pairwise, union, medoid, outlier (required).-n- number of clusters (required).-i- similarity index, RR or SM (required).-t- Fraction of outliers to trim in decimals (default is None).-w- Weighing clusters by frames it contains (default is True).-d- directory where thenormed_clusttraj.c*.npyfiles are located (required)-s- location wheresummaryfile is located with population of each cluster (required)
python ../../utils/similarity.py -m union -n 10 -i SM -t 0.1 -d ../normalization -s ../clusters/outputs/summary_20.txtTo generate a similarity dictionary using data in ../normalization (make sure you are in the prime directory) using the union method (2.2 in Fig 2) and Sokal Michener index. In addition, 10% of the outliers were trimmed. You can either python exec_similarity.py or run example above.
The result is a dictionary organized as followes: Keys are frame #. Values are [cluster 1 similarity, cluster #2 similarity, ..., average similarity of all clusters].
scripts/prime/exec_rep_frames.py will determine the native structure of the protein using the similarity dictionary generated in step 4.
-h - for help with the argument options.
-m - methods (for one method, None for all methods)
-s - folder to access for w_union_SM_t10.txt file
-i - similarity index (required)
-t - Fraction of outliers to trim in decimals (default is None).
-d - directory where the normed_clusttraj.c* files are located (required if method is None)
python ../../utils/rep_frames.py -m union -s outputs -d ../normalization -t 0.1 -i SMFor more information on the PRIME algorithm, please refer to the PRIME paper. Please cite using CITATION.bib.
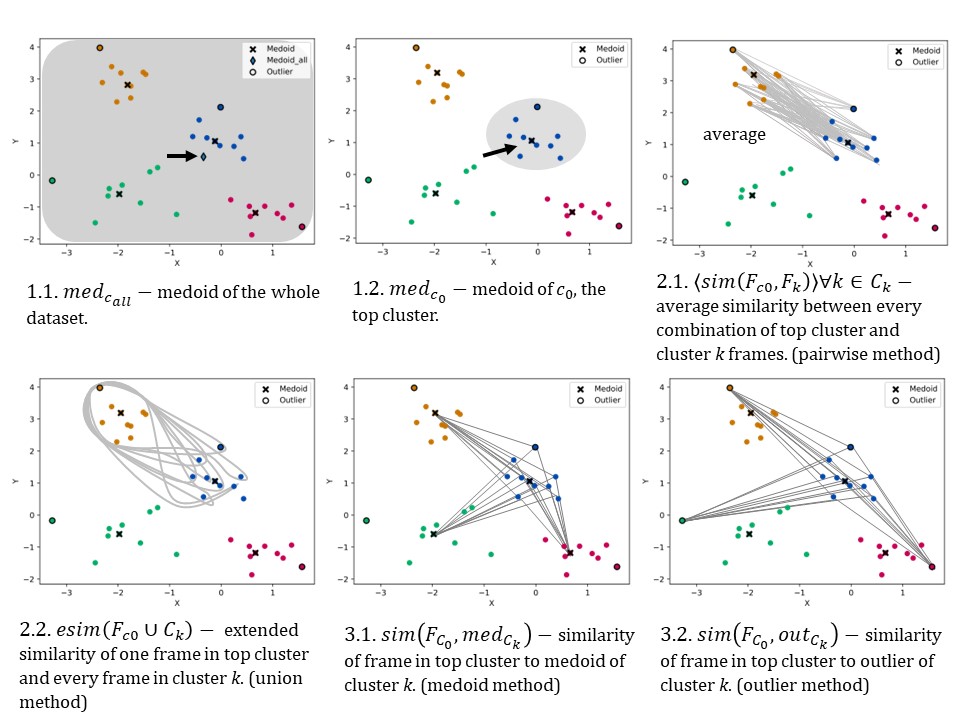
Fig 2. Six techniques of protein refinement. Blue is top cluster.