NanoCube is a minimalistic in-memory, in-process OLAP engine for lightning fast point queries on Pandas DataFrames. As of now, less than 50 lines of code are required to transform a Pandas DataFrame into a multi-dimensional OLAP cube. NanoCube shines when point queries need to be executed on a DataFrame, e.g. for financial data analysis, business intelligence or fast web services.
If you think it would be valuable to extend NanoCube with additional OLAP features please let me know. You can reach out by opening an issue or contacting me on LinkedIn.
pip install nanocubeimport pandas as pd
from nanocube import NanoCube
# create a DataFrame
df = pd.read_csv('sale_data.csv')
value = df.loc[(df['make'].isin(['Audi', 'BMW']) & (df['engine'] == 'hybrid')]['revenue'].sum()
# create a NanoCube and run sum aggregated point queries
# Declare the column supposed to be aggregated in `measures` and filtered in `dimensions`
nc = NanoCube(df, dimensions=["make", "engine"], measures=["revenue"])
for i in range(1000):
value = nc.get('revenue', make=['Audi', 'BMW'], engine='hybrid')Tip: Only include those columns in the NanoCube setup, that you actually want to query! The more columns you include, the more memory and time is needed for initialization.
df = pd.read_csv('dataframe_with_100_columns.csv') nc = NanoCube(df, dimensions=['col1', 'col2'], measures=['col100'])
Tip: Use dimensions with highest cardinality first. This yields much faster response time when more than 2 dimensions need to be filtered.
nc.get(promo=True, discount=True, customer='4711') # bad=slower, non-selevtive columns first nc.get(customer='4711', promo=True, discount=True) # good=faster, most selective column first
For aggregated point queries NanoCube are up to 100x or even 1,000x times faster than Pandas. When proper sorting is applied to your DataFrame, the performance might improve even further.
For the special purpose of aggregative point queries, NanoCube is even by factors faster than other DataFrame oriented libraries, like Spark, Polars, Modin, Dask or Vaex. If such libraries are a drop-in replacements for Pandas, then you should be able to accelerate them with NanoCube too. Try it and let me know.
NanoCube is beneficial only if some point queries (> 5) need to be executed, as the initialization time for the NanoCube needs to be taken into consideration. The more point query you run, the more you benefit from NanoCube.
NanoCube creates an in-memory multi-dimensional index over all relevant entities/columns in a dataframe. Internally, Roaring Bitmaps (https://roaringbitmap.org) are used for representing the index. Initialization may take some time, but yields very fast filtering and point queries.
Approach: For each unique value in all relevant dimension columns, a bitmap is created that represents the rows in the DataFrame where this value occurs. The bitmaps can then be combined or intersected to determine the rows relevant for a specific filter or point query. Once the relevant rows are determined, Numpy is used then for to aggregate the requested measures.
NanoCube is a by-product of the CubedPandas project (https://github.com/Zeutschler/cubedpandas) and will be integrated into CubedPandas in the future. But for now, NanoCube is a standalone library that can be used with any Pandas DataFrame for the special purpose of point queries.
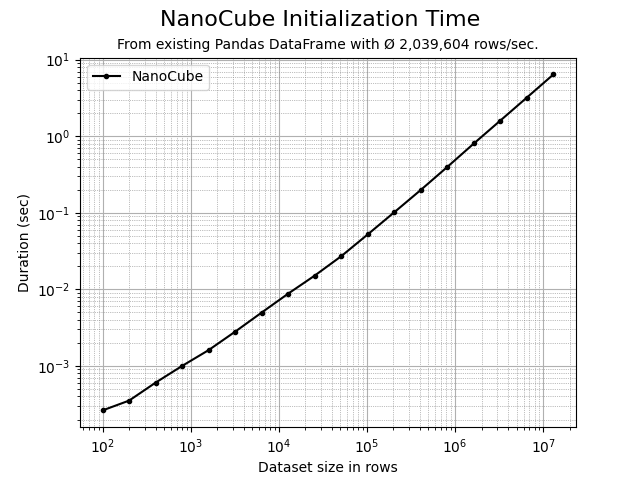
NanoCube is free and MIT licensed. The prices to pay are additional memory consumption, depending on the use case typically 25% on top of the original DataFrame and the time needed for initializing the multi-dimensional index, typically 250k rows/sec depending on the number of columns to be indexed and your hardware. The initialization time is proportional to the number of rows in the DataFrame (see below).
You may want to try and adapt the included samples sample.py and benchmarks
benchmark.py and benchmark.ipynb to test the behavior of NanoCube
on your data.
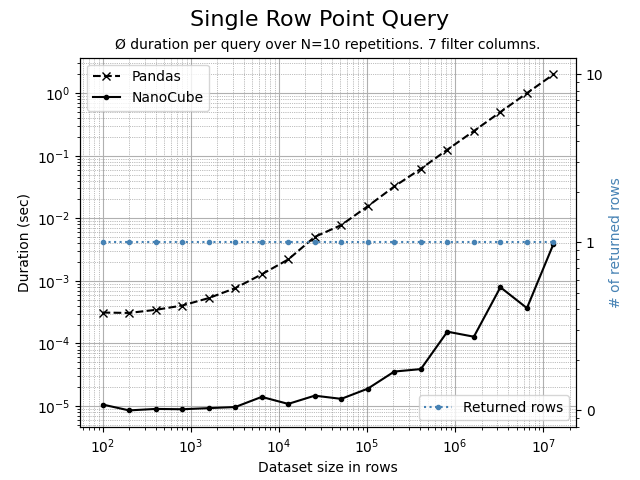
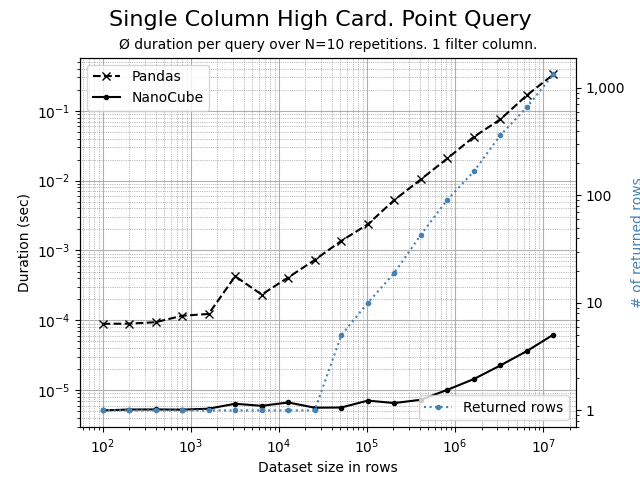
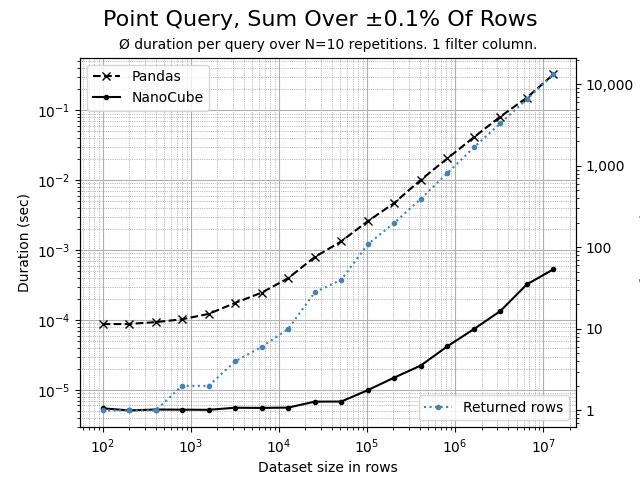
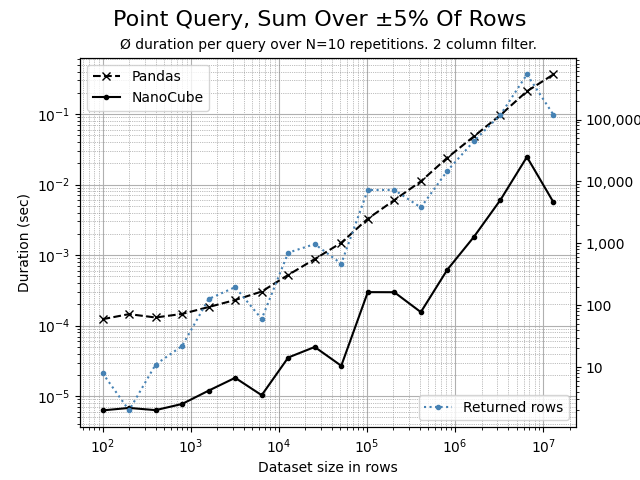
Using the Python script benchmark.py, the following comparison charts can be created. The data set contains 7 dimension columns and 2 measure columns.
A highly selective query, fully qualified and filtering on all 7 dimensions. The query will return and aggregates 1 single row. NanoCube is 100x or more times faster than Pandas.
If sorting is applied to the DataFrame - low cardinality dimension columns first, higher dimension cardinality columns last - then the performance of NanoCube can potentially improve dramatically, ranging from 1.1x up to ±10x or even 100x times. Here, the same query as above, but the DataFrame was sorted beforehand.
A highly selective, filtering on a single high cardinality dimension, where each member represents ±0.01% of rows. NanoCube is 100x or more times faster than Pandas.
A highly selective, filtering on 1 dimension that affects and aggregates 0.1% of rows. NanoCube is 100x or more times faster than Pandas.
A barely selective, filtering on 2 dimensions that affects and aggregates 5% of rows. NanoCube is consistently 10x faster than Pandas. But you can already see, that the aggregation in Numpy become slightly more dominant.
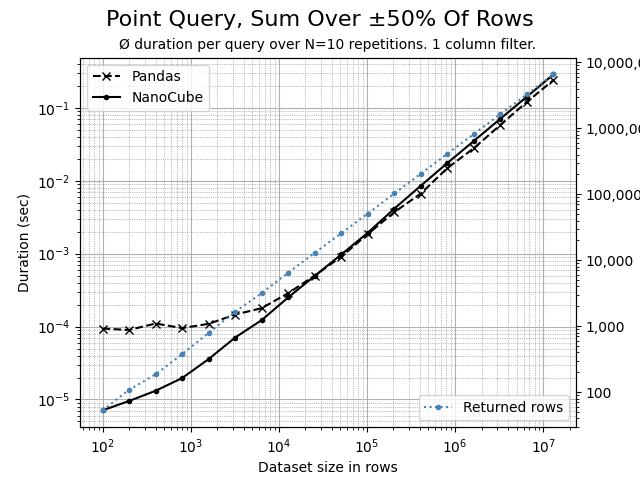
A non-selective query, filtering on 1 dimension that affects and aggregates 50% of rows. Here, most of the time is spent in Numpy, aggregating the rows. The more rows, the closer Pandas and NanoCube get as both rely on Numpy for aggregation.
The time required to initialize a NanoCube instance is almost linear. The initialization throughput heavily depends on the number of dimension columns. A custom file format will be added soon allowing ±4x times faster loading of a NanoCube in comparison to loading the respective parquet dataframe file using Arrow.