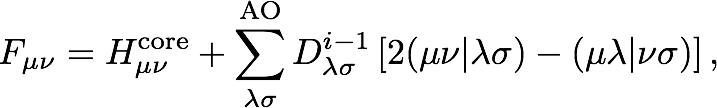The use of point-group symmetry in quantum chemical calculations can lead to exceptionally efficient programs, both in terms of their memory/disk usage and their computing times. If a molecule has even one element of symmetry, for example, this can reduce the number of wave function parameters that must be stored by up to a factor of two (the order of the point group) and the computing effort by up to a factor of four (the square of the order of the point group). Most quantum chemical programs can take advantage of Abelian symmetry -- more accurately, D2h and its subgroups -- which means that higher symmetry structures can yield up to a factor of 64 reduction in the computational costs.
The purpose of this project is to take advantage of basic point-group symmetry in an SCF calculation. While the most efficient programs will store only the symmetry-allowed sets of two-electron integrals, for the purposes of this project we'll deal only with the one-electron (two-index) components.
The vanishing integral rule of point-group theory may be stated as follows for Abelian groups:
where ΓX denotes the irreducible representation (irrep) of entity X. That is, unless the direct product of the irreps of the functions or operators in the integrand contains the totally symmetric irrep, the integral must be zero.
The vanishing integral rule of group theory may be readily applied to the various integrals and wave-function parameters in the SCF procedure if the AO-basis functions over which the they are evaluated have been properly symmetrized [i.e. used to form symmetry-adapted linear combinations, normally referred to as symmetry orbitals (SOs)].
For example, consider the H2O test case with the STO-3G basis set as given in Project #3. The AO (contracted Gaussian) basis functions on the oxygen atom consist of 1s, 2s, 2px, 2py, and 2pz orbitals, which those for each hydrogen atom consist of only the 1s, giving a total of seven AOs. If we take oxygen to be the first atom and ignore the C2v point group symmetry of the molecule, the resulting overlap integral matrix looks like:
1 2 3 4 5 6 7
1 1.0000000 0.2367039 -0.0000000 -0.0000000 -0.0000000 0.0384056 0.0384056
2 0.2367039 1.0000000 0.0000000 -0.0000000 0.0000000 0.3861388 0.3861388
3 -0.0000000 0.0000000 1.0000000 -0.0000000 0.0000000 0.2684382 -0.2684382
4 -0.0000000 -0.0000000 -0.0000000 1.0000000 -0.0000000 0.2097269 0.2097269
5 -0.0000000 0.0000000 0.0000000 -0.0000000 1.0000000 -0.0000000 -0.0000000
6 0.0384056 0.3861388 0.2684382 0.2097269 -0.0000000 1.0000000 0.1817599
7 0.0384056 0.3861388 -0.2684382 0.2097269 -0.0000000 0.1817599 1.0000000
The first five row/column indices correspond to the basis functions on the oxygen (in the order listed above), while rows/columns six and seven correspond to the basis functions on each hydrogen. Thus, the 1-2 element is the overlap integral between the 1s and 2s orbitals on the oxygen, while element 3-7 is the overlap integral between the oxygen 2px orbital and the second hydrogen 1s orbital.
Points to note about this matrix:
- The 1s and 2s orbitals on oxygen are not orthogonal to one another, but both are orthogonal to all three p-type orbitals. This occurs because, in Cartesian Gaussian basis sets, orthogonality is generally obtained only between functions of different angular momenta on the same atomic center.
- The 1s orbitals on the hydrogens have the same overlap integral (to within a sign) with all the orbitals on the oxygen because they are symmetry equivalent. However, the 1s orbitals alone do not transform as irreps of the C2v point group.
- The hydrogen 1s orbitals are orthogonal to the 2pz orbital on the oxygen (cf. elements 5-6 and 5-7). This is because the water molecule was arbitrarily oriented in the xy-plane to compute the above integrals; thus the 2pz orbital is perpendicular to the molecular plane containing the hydrogen 1s orbitals.
What happens if we turn on symmetry, i.e. take linear combinations of the AO to obtain SOs? If we orient the molecule oriented in the yz-plane, so that the z-axis corresponds to the C2 rotation axis, we find that the AOs transform as the following irreps:
| SO | Irrep |
|---|---|
| O 1s | A1 |
| O 2s | A1 |
| O 2px | B1 |
| O 2py | B2 |
| O 2pz | A1 |
| H1 1s + H2 1s | A1 |
| H1 1s - H2 1s | B2 |
Thus, after symmetrization, there are four A1 SOs, one B1 SO, and two B2 SOs. (The STO-3G basis set contains no A2 orbitals. One must include at least d-type functions on the oxygen and/or p-type functions on the hydrogens to obtain A2 orbitals.) Note that a normalization factor of 1/sqrt(2) should be included for the H 1s SOs above.
In the SO basis, the overlap integral matrix becomes:
1 2 3 4 5 6 7
1 1.0000000 0.2367039 -0.0000000 0.0543137 0.0000000 0.0000000 0.0000000
2 0.2367039 1.0000000 -0.0000000 0.5460828 0.0000000 0.0000000 0.0000000
3 -0.0000000 -0.0000000 1.0000000 0.2965987 0.0000000 0.0000000 0.0000000
4 0.0543137 0.5460828 0.2965987 1.1817599 0.0000000 0.0000000 0.0000000
5 0.0000000 0.0000000 0.0000000 0.0000000 1.0000000 0.0000000 0.0000000
6 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.0000000 0.3796290
7 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.3796290 0.8182401
Points to note about this matrix:
- The matrix now has a block-diagonal structure, with 4x4, 1x1, and 2x2 sub-matrices, corresponding to the A1, B1, and B2 irreps, respectively. All off-diagonal blocks correspond to overlaps between SOs of different irreps, which must be zero according to the vanishing integral rule.
- There is only one B1 SO (the O 2px orbital), and it is normalized.
- The oxygen 1s and 2s orbitals are still orthogonal to the oxygen 2pz because they are functions of different angular momentum on the same atomic center.
- The same block-diagonal structure will appear for the kinetic-energy (T), one-electron potential-energy (V), and core Hamiltonian (H) integrals.
The PSI checkpoint file contains a great deal of useful symmetry information. To convert your SCF program to make use of symmetry, two key pieces of information are:
- The number of irreps in the molecular point group. Use the
chkpt_rd_nirreps()function to retrieve this integer. - The number of SOs per irrep, available as an integer array from the
chkpt_rd_sopi()function.
The block-diagonal structure above implies that one need only store and use the sub-matrices of the integrals rather than the full matrix. One way of doing this is to store the one-electron integrals as an array of smaller matrices, the size of which is dictated by the number of SOs per irrep. For example, here's a code snippet that will extract the one-electron integrals from the appropriate file (as was done in Project #7 ), but stores them in an array of matrices:
int ntri, nirreps;
double *scratch;
double ***S;
...
ntri = nso*(nso+1)/2;
scratch = init_array(ntri);
...
symm_offset = init_int_array(nirreps);
for(h=1; h < nirreps; h++) symm_offset[h] = symm_offset[h-1] + sopi[h-1];
...
/* overlap integrals */
S = (double ***) malloc(nirreps*sizeof(double **));
stat = iwl_rdone(PSIF_OEI, PSIF_SO_S, scratch, ntri, 0, 0, outfile);
for(h=0; h < nirreps; h++) {
S[h] = block_matrix(sopi[h], sopi[h]);
for(i=0; i < sopi[h]; i++)
for(j=0; j <= i; j++) {
ij = INDEX(i+symm_offset[h],j+symm_offset[h]);
S[h][i][j] = S[h][j][i] = scratch[ij];
}
}Notice how S is now a double *** -- i.e. an array of matrices. This code uses the same INDEX() function found in
Project #3,
and the sopi array is the integer array of SOs per irrep obtained using the chkpt_rd_sopi() function.
The symm_offset array is useful for converting between relative orbital indices and absolute orbital indices.
(For example, orbital 0 in the B2 irrep block corresponds to orbital 5
in the full list of orbitals; the first number is the relative index, the second is the absolute index.)
If we store the integral, MO coefficient, density, and Fock matrices in the block-diagonal structure described above, we may compute matrix products of these quantities separately for each diagonal block. For example, the matrix product may be written as:
where h denotes a particular irrep of the point group and Ch denotes the h-th irrep subblock of the full matrix C. (The notation of the large plus with a circle around it indicates a direct sum.)
The calculation of the density matrix requires information about the number of occupied orbitals:
In a calculation without symmetry, one need only know the number of electrons (equivalently, the atomic numbers of the atoms and the overall molecular charge) to evaluate this expression. However, if the density and MO coefficient matrices are stored in symmetry-blocked form, as above, one needs to know the number of occupied MOs in each irrep.
For the case of the C2v water molecule, which has two σ O-H bonds, two oxygen lone pairs, and one oxygen core orbital, the orbital occupations are not difficult to determine, given the above information about the number of SOs in each irrep. Since the MOs are constructed as linear combinations of the SOs, and only SOs of the same irrep can combine, we may rationalize the occupied orbitals of H2O as follows:
| Occupied MO | Irrep | Description |
|---|---|---|
| O 1s | A1 | Oxygen Core |
| O 2s + (H1 1s + H2 1s) | A1 | Symmetric Combination of O-H σ Bonds |
| O 2pz | A1 | In-Plane Oxygen Lone Pair |
| O 2px | B1 | Out-of-Plane Oxygen Lone Pair |
| O 2py + (H1 1s - H2 1s) | B2 | Antisymmetric Combination of O-H σ Bonds |
Thus, for the water molecule, there are 3 A1, 1 B1, and 1 B2 occupied orbitals.
Determining the occupations automatically is sometimes a difficult task, especially for molecules with unusual bonding patterns. Many SCF programs will simply take the core Hamiltonian matrix (often used as the initial guess for the Fock matrix), diagonalize it (in the othogonal AO basis) in each irrep block, and identify the lowest eigenvalues in each irrep as occupied. However, this approach often fails for many molecules (especially those with open shells) because of near-degeneracies that may not be adequately described by the simple core Hamiltonian. Thus, some programs will make use of more sophisticated guesses at the Fock matrix, e.g. Hueckel orbitals or other (semi)empirical forms.
The only portion of the SCF procedure involving two-electron integrals is the Fock-matrix build:
where the notation used in Project #3 is retained above. If the Fock matrix and density matrix are stored in a symmetry-blocked manner similar to the overlap matrix above, then one may limit the corresponding loops only to the symmetry-allowed combinations, e.g. the irreps of μ and
ν must match for Fμν to be non-zero and the irreps of λ and σ must match for Dλσ to be non-zero. Note, however, that, because of the symmetries in the two-electron integrals, the irrep of λ and σ do not necessarily have to match that of μ and ν.- Hint: Fock-Build Code