This inductive link prediction competition accompanies the KG Course and welcomes students' attempts to improve the link prediction performance on two newly constructed datasets.
This repo contains:
- The datasets in the
./datafolder - A boilerplate code with 2 baselines that you can base your implementations on
The code employs the PyKEEN framework for training KG link prediction models.
Main requirements:
- python >= 3.9
- torch >= 1.10
You will need PyKEEN 1.8.0 or newer.
$ pip install pykeenBy the time of creation of this repo 1.8.0 is not yet there, but the latest version from sources contains everything we need
$ pip install git+https://github.com/pykeen/pykeen.gitIf you plan to use GNNs (including the InductiveNodePieceGNN baseline) make sure you install torch-scatter
and torch-geometric
compatible with your python, torch, and CUDA versions.
Running the code on a GPU is strongly recommended.
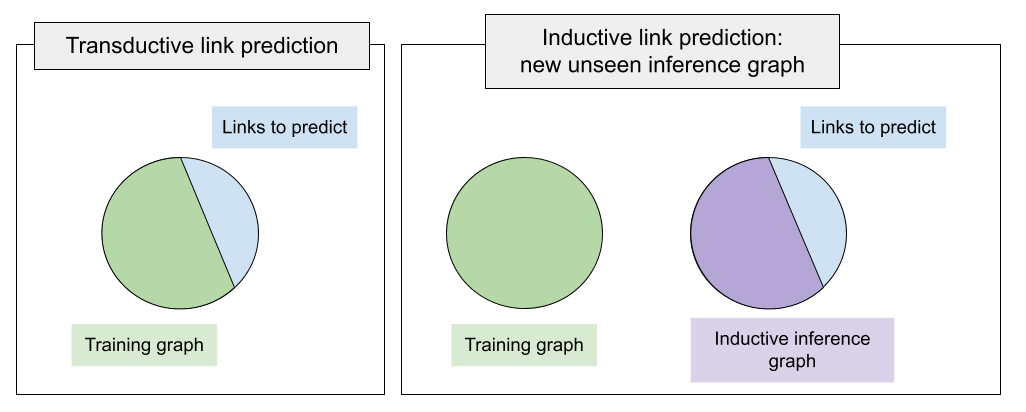
Inductive link prediction is different from the standard transductive task in a way that at inference time you are given a new, unseen graph with unseen entities (but known relation types). Here is the schematic description of the task:
Here, we provide 2 inductive LP datasets. Each dataset in ./data consists of 4 splits:
train.txt- the training graph on which you are supposed to train a modelinference.txt- the inductive inference graph disjoint with the training one - that is, it has a new non-overlapping set of entities, the missing links are sampled from this graphinductive_validation.txt- validation set of triples to predict, uses entities from the inference graphinductive_test.txt- test set of triples to predict, uses entities from the inference graph- a held-out test set of triples - kept by the organizers for the final ranking 😉 , uses entities from the inference graph
small dataset stats:
| Split | Entities | Relations | Triples |
|---|---|---|---|
| Train | 10,230 | 96 | 78,616 |
| Inference | 6,653 | 96 (subset) | 20,960 |
| Inference validation | 6,653 | 96 (subset) | 2,908 |
| Inference test | 6,653 | 96 (subset) | 2,902 |
| Held-out test set | 6,653 | 96 (subset) | 2,894 |
large dataset stats:
| Split | Entities | Relations | Triples |
|---|---|---|---|
| Train | 46,626 | 130 | 202,446 |
| Inference | 29,246 | 130 (subset) | 77,044 |
| Inference validation | 29,246 | 130 (subset) | 10,179 |
| Inference test | 29,246 | 130 (subset) | 10,184 |
| Held-out test set | 29,246 | 130 (subset) | 10,172 |
Training shallow entity embeddings in this setup is useless as trained embeddings cannot be used for inference over unseen entities. That's why we need new representation learning mechanisms - in particular, we use NodePiece for the baselines.
NodePiece in the inductive mode will use the set of relations seen in the training graph to tokenize entities in the training and inference graphs. We can afford tokenizing the nodes in the inference graph since the set of relations is shared between training and inference graphs (more formally, the set of relations of the inference graph is a subset of training ones).
We offer here 2 baselines:
InductiveNodePiece- plain tokenizer + tokens MLP encoder to bootstrap node representations. Fast.InductiveNodePieceGNN- everything above + an additional 2-layer CompGCN message passing encoder. Slower but attains higher performance.
For more information on the models check out the PyKEEN tutorial on inductive link prediction with NodePiece
Both baselines are implemented in the main.py.
CLI arguments:
Usage: main.py [OPTIONS]
Options:
-ds, --dataset_size TEXT # "small" or "large"
-dim, --embedding_dim INTEGER
-tokens, --tokens_per_node INTEGER # for NodePiece
-lr, --learning_rate FLOAT
-m, --margin FLOAT # for the margin loss and SLCWA training
-negs, --num_negatives INTEGER # negative samples per positive in the SLCWA regime
-b, --batch_size INTEGER
-e, --num_epochs INTEGER
-wandb, --wandb BOOLEAN
-save, --save_model BOOLEAN
-gnn, --gnn BOOLEAN # for activating InductiveNodePieceGNNEvaluation metrics (more documentation):
- MRR (Inverse Harmonic Mean Rank) - higher is better
- Hits @ 100 - higher is better
- Hits @ 10
- Hits @ 5
- Hits @ 3
- Hits @ 1
- MR (Mean Rank) - lower is better
- Adjusted Arithmetic Mean Rank (AMR) - lower in better
| Model | MRR | H@100 | H@10 | H@5 | H@3 | H@1 | MR | AMR |
|---|---|---|---|---|---|---|---|---|
| InductiveNodePieceGNN | 0.1326 | 0.4705 | 0.2509 | 0.1899 | 0.1396 | 0.0763 | 881 | 0.270 |
| InductiveNodePiece | 0.0381 | 0.4678 | 0.0917 | 0.0500 | 0.0219 | 0.007 | 1088 | 0.334 |
Configs:
- InductiveNodePieceGNN (32d, 50 epochs, 24K params) - NodePiece (5 tokens per node, MLP aggregator) + 2-layer CompGCN with DistMult composition function + DistMult decoder. Training time: 77 min*
main.py -dim 32 -e 50 -negs 16 -m 2.0 -lr 0.0001 --gnn True- InductiveNodePiece (32d, 50 epochs, 15.5K params) - NodePiece (5 tokens per node, MLP aggregator) + DistMult decoder. Training time: 6 min*
main.py -dim 32 -e 50 -negs 16 -m 5.0 -lr 0.0001| Model | MRR | H@100 | H@10 | H@5 | H@3 | H@1 | MR | AMR |
|---|---|---|---|---|---|---|---|---|
| InductiveNodePieceGNN | 0.0705 | 0.374 | 0.1458 | 0.0990 | 0.0730 | 0.0319 | 4566 | 0.318 |
| InductiveNodePiece | 0.0651 | 0.287 | 0.1246 | 0.0809 | 0.0542 | 0.0373 | 5078 | 0.354 |
Configs:
- InductiveNodePieceGNN (32d, 53 epochs, 24K params) - NodePiece (5 tokens per node, MLP aggregator) + 2-layer CompGCN with DistMult composition function + DistMult decoder. Training time: 8 hours*
main.py -dim 32 -e 53 -negs 16 -m 20.0 -lr 0.0001 -ds large --gnn True- InductiveNodePiece (32d, 17 epochs, 15.5K params) - NodePiece (5 tokens per node, MLP aggregator) + DistMult decoder. Training time: 5 min*
main.py -dim 32 -e 17 -negs 16 -m 15.0 -lr 0.0001 -ds large* Note: All models were trained on a single RTX 8000. Average memory consumption during training is about 2 GB VRAM on the small dataset and about 3 GB on large.
- Fork the repo
- Train your inductive link prediction model
- Save the model weights using the
--save Trueflag - Upload model weights on GitHub or other platforms (Dropbox, Google Drive, etc)
- Open an issue in this repo with the link to your repo, performance metrics, and model weights