The goal of my_project is to provide a minimal infrastructure for writing a research paper using R packages targets as the main build automation data analysis pipeline and bookdown for authoring the manuscript.
-
Fork this project to your Github account
-
Create a new R project (in RStudio) from Version Control
2.1 Choose a directory and paste the https url of your Github repo
-
Restore the development environment with
renv::restore() -
In Tools -> Project Options -> Build tools, select Website and define your
my_project/Articleas the site directory
4.1 The Build pane will be available, from where you can build your article (Ctrl+Shif+B) when your pipeline is up to date
After this initial setup your workflow would consist of creating new functions to perform a task, declare a target for such a task, run the pipeline with targets::tar_make(), update your manuscript in the Article folder and, compile the article (Build Book or Ctrl+Shif+B). Your can check your workflow using the targets functions tar_visnetwork() or tar_manifest(). For more advanced usage you should check the targets manual. The targets dependency graph for this project is presented in the figure bellow.
The data analysis pipeline is used to produce artifacts (e.g. intermediary data, data frames, regression results, etc.), LaTEX tables (i.e., .tex files) and figures, which can be consumed in the article source RMarkdown. For example, the pipeline would create a LaTEX table file in the directory Article/Tables named hello.tex and, the index.Rmd in the Article folder, which will compile into the paper PDF, would call \input{Tables/hello} to insert the table into the final document.
The folder and files structure is chosen to benefit from the data analysis pipeline tool, the targets package and, the writting of a research article using bookdown.
|--- Article/ : where the files of your article will be
|------ _bookdown.yml : bookdown configuration of your article
|------ _output.yml : output formats compatible with bookdown
|------ article/ : corresponds to the output_dir option in _bookdown.yml
|------ Figs/ : figures created in the pipeline should be saved here
|------ index.Rmd : the first Rmd file of your article. THIS IS NOT a Target Markdown file
|------ preamble.tex : custom preamble to be inserted into your article TEX file
|------ Tables/ : LaTEX tables generated in the pipeline should be saved here
|------ references.bib : your BIB file
|--- Data/ : folder where all your data is saved for ingestion
|----- input_data.csv
|--- Presentation/
|------ Figs/ : figures created for presentation slides in the pipeline should be saved here
|------ Tables/ : LaTEX tables generated for presentation slides in the pipeline should be saved here
|--- R/ : folder of scripts with user-defined R code (functions)
|----- functions_data.R : data pre-processing functions
|----- functions_analysis.R : data analysis (i.e., regressions, ML models)
|----- functions_tables.R : generating LaTEX tables from results
|----- functions_plots.R : functions creating figures
|--- renv/ : dependency management for improved reproducibility
|--- _targets/ : where tar_make() writes target storage and metadata
|--- LICENSE.md
|--- README.md
|--- renv.lock
|--- _targets.R : target script file
I have opted to keep separate workflows, data analysis using targets and, article authoring using bookdown and the Build pane, mainly because tarchetypes::tar_render() does not support a bookdown::pdf_book: with many files (e.g., one file for each paper section). Another good reason not to put the article compilation into the pipeline is the time it takes to complete the task and, we usually do not want to recompile the whole article every single time we create or adjust an intermediary result. Keeping the data analyis apart from the article authoring lets us interactively make our analysis and debug results without worrying about the article's format and other details. Only when we are happy with our analysis, we can put everything together in the manuscript and build it (CTRL+SHIFT+B) to check on the result.
Of course, we still have the ability to introduce the compilation of an RMarkdown file into the pipeline, see the section on Literate Programming from the targets documentation. In this case, we rather compile a lightweight report which depends on other targets in the pipeline. Those targets are usually loaded through functions like tar_load() and used throughout the report. Unless your research article falls into the lightweight category, I suggest you keep the article's build process and the data analysis apart.
The following are general guidelines for using R on research projects. This section is heavily inspired on R_guide from professor Sean Higgins, all credit goes to him.
-
Use
data.table. The efficiency advantages will payoff. The learning curve may be steeper thantidyversebut, the computation speed and lower memory footprint are well worth it. -
Use
assertthat::assert_that()frequently to add programmatic sanity checks in the code. -
Use pipes. Either the R-native |> or, the
magrittr's%>%. -
Use
fixestfor all your regressions. It's fast, handles fixed-effects neatly, large datasets and, multiple regression specifications at once. -
Use
modelsummaryto create the tables fromfixestregressions. If needed, use eitherkableExtraorgtto further format them. For LaTEX output only, you can also use thefixest::etablefunction.
-
Use
ggplot2. This is a no-brainer. -
Use customized themes and colorblind-friendly palettes. The package
ggthemes,ggpubrand many others are your friends. In theRfolder you will find a customized theme namedtheme_academicin theutils.Rfile. -
For maps, use the
sfpackage. This package makes plotting maps easy (with ggplot2::geom_sf()), and also makes other tasks like joining geocoordinate polygons and points a breeze. -
Save your ggplots with
ggplot2::ggsave(). For reproducible graphs, always specify the width and height arguments inggplot2::ggsave().
-
Read csv files with
data.table::fread(). -
Smaller datasets can be saved as csv files.
data.table::fwrite()does the job. -
Larger datasets should be saved using
fstpackage'sfst::write_fst(). If serialization is not possible (e.g., due to the presence of list-columns), usesaveRDS().
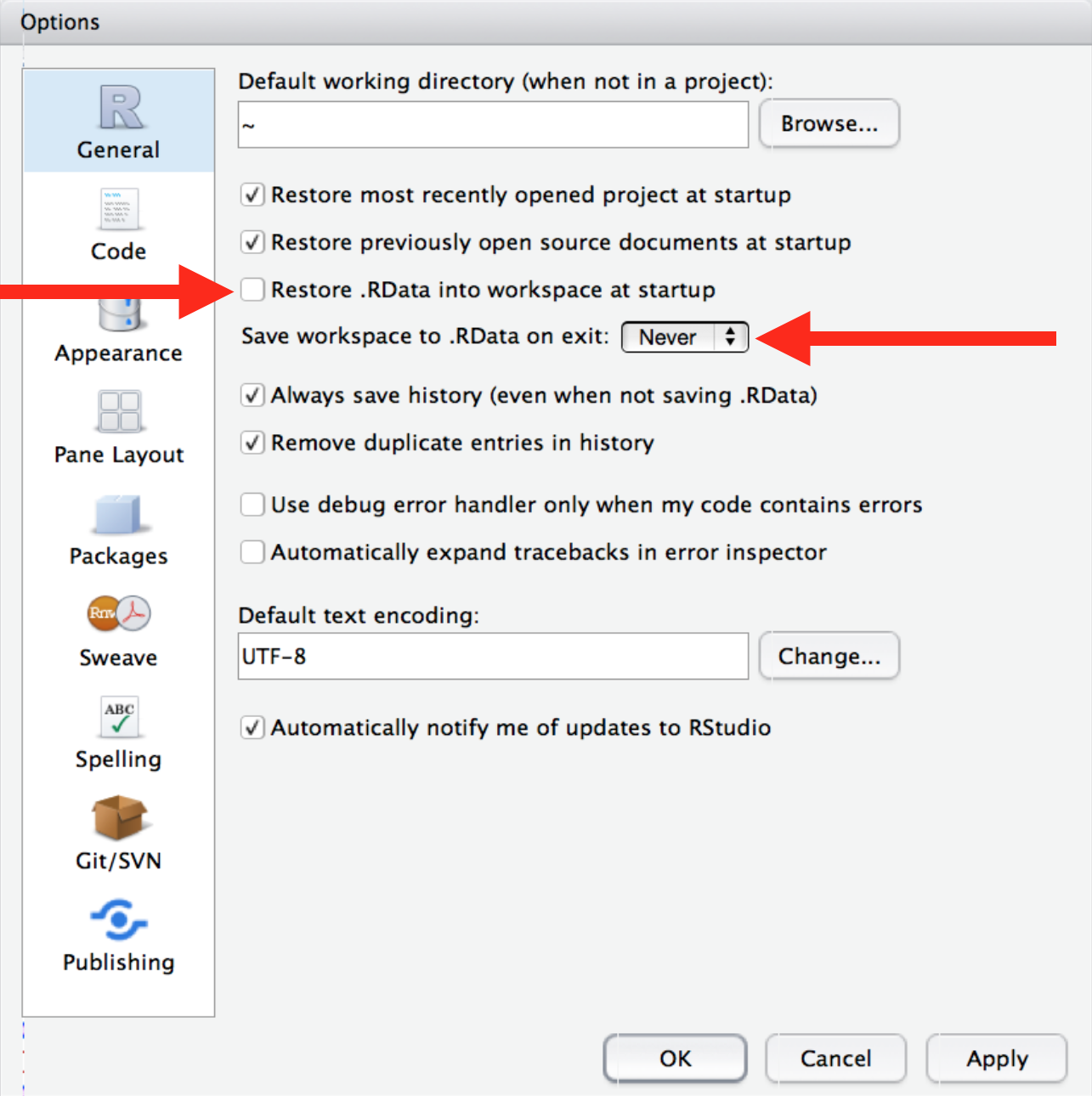
Reproducibility matters. You want other researchers (including future you!) to be able to replicate your results. Never save the workspace when quitting RStudio and, do not reload it from an .Rdata file. Go to Tools > Global Options > General and:
-
Never use
setwd()or absolute file paths. Instead, use relative file paths with theherepackage. Did I say never? Never. -
Lear how to use RStudio Projects and keep all project-related files under one working directory.
-
Use
renvto manage the packages used in an RStudio project, avoiding conflicts related to package versioning. -
USE a version control system, I suggest Git/Github. I cannot emphasize this enough, if you want to control all changes made during development of your piece of software and many versions of it, learn how to use Git! Suggested places to start, Grant Mcdermott's slides, Happy Git and GitHub for the useR and, about Git.
-
Adopt a commit messages convention. Be it the conventional commits or angular, adhere to one specification and then use an automatic changelog generator like
git-changelog. -
Commit frequently. Ideally, there would be one commit for each specific change made in the code.