TL;DR: You just need a
dataset pointand amodel prediction function. CFNOW will find the closest point with a different class.
The simplest way to generate counterfactuals for any tabular dataset and model.
This package finds an optimal point (closer to the input dataset point), which the classification is different from the original classification (i.e. "flips" the classification of the original input by minimally changin it).
- Minimal example
- Counterfactual Charts
- I have: binary categorical features!
- I have: one-hot encoded features!
- I have: binary and OHE features!
- How to cite
- Python >= 3.8
from cfnow import find_tabular
import sklearn.datasets
import sklearn.ensemble
import pandas as pd
# Generating a sample model
X, y = sklearn.datasets.load_iris(return_X_y=True)
model = sklearn.ensemble.RandomForestClassifier()
model.fit(X, y)
# Selecting a random point
x = X[0]
# Here we can see the original class
print(f"Factual: {x}\nFactual class: {model.predict([x])}")
# Then, we use CFNOW to generate the minimum modification to change the classification
cf_obj = find_tabular(
factual=pd.Series(x),
model_predict_proba=model.predict_proba,
limit_seconds=10)
# Here we can see the new class
print(f"CF: {cf_obj.cfs[0]}\nCF class: {model.predict([cf_obj.cfs[0]])}")This package is integrated with CounterPlots, that allows you to graphically represent your counterfactual explanations!
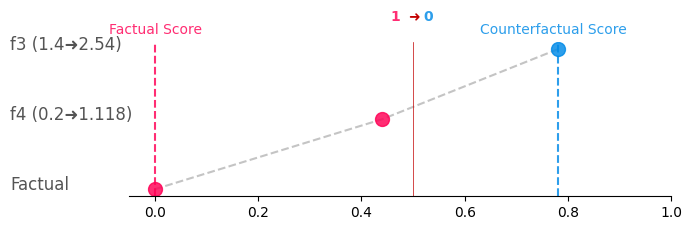
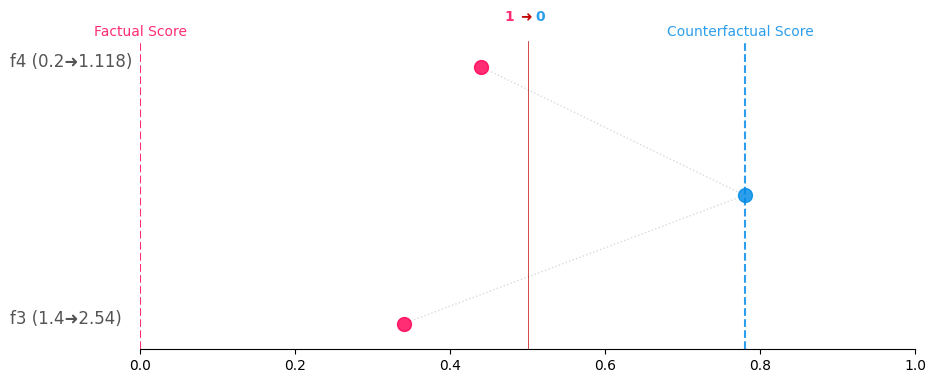
You can simply generate Greedy, CounterShapley, and Constellation charts for a given CF with:
# Get the counterplots for the first CF and save as greedy.png
cf_obj.generate_counterplots(0).greedy('greedy.png')# Get the counterplots for the first CF and save as greedy.png
cf_obj.generate_counterplots(0).countershapley('countershapley.png')# Get the counterplots for the first CF and save as greedy.png
cf_obj.generate_counterplots(0).constellation('constellation.png')The minimal example above considers all features as numerical continuous, however, some datasets can have categorical (binary or one-hot encoded) features. CFNOW can handle these data types in a simple way as demonstrated below:
import pandas as pd
import numpy as np
import sklearn.ensemble
# Generate data with 5 binary categorical features and 3 continuous numerical features
X = np.hstack((
np.random.randint(0, 2, size=(1000, 5)),
np.random.rand(1000, 3) * 100
))
# Random binary target variable
y = np.random.randint(0, 2, 1000)
# Train RandomForestClassifier
model = sklearn.ensemble.RandomForestClassifier().fit(X, y)
# Display the original class for a random test sample
x = X[0]
print(f"Factual: {x}\nFactual class: {model.predict([x])}")from cfnow import find_tabular
# Then, we use CFNOW to generate the minimum modification to change the classification
cf_obj = find_tabular(
factual=pd.Series(x),
feat_types={i: 'cat' if i < 5 else 'cont' for i in range(8)},
model_predict_proba=model.predict_proba,
limit_seconds=10)
# Here we can see the new class
print(f"CF: {cf_obj.cfs[0]}\nCF class: {model.predict([cf_obj.cfs[0]])}")
import warnings
import pandas as pd
import numpy as np
from sklearn.preprocessing import OneHotEncoder
import sklearn.ensemble
warnings.filterwarnings("ignore", message="X does not have valid feature names, but RandomForestClassifier was fitted with feature names")
# Generate data
X = np.hstack((np.random.randint(0, 10, size=(1000, 5)), np.random.rand(1000, 3) * 100))
# One-hot encode the first 5 categorical columns
# !!!IMPORTANT!!! The naming of OHE encoding features columns MUST follow feature_value format.
# Therefore, for a feature called color with value equal to red or blue, the OHE encoding columns
# must be named color_red and color_blue. Otherwise, the CF will not be able to find the correct
# columns to modify.
encoder = OneHotEncoder(sparse=False)
X_cat_encoded = encoder.fit_transform(X[:, :5])
names = encoder.get_feature_names_out(['cat1', 'cat2', 'cat3', 'cat4', 'cat5'])
# Combine and convert to DataFrame
df = pd.DataFrame(np.hstack((X_cat_encoded, X[:, 5:])), columns=list(names) + ['num1', 'num2', 'num3'])
# Random binary target variable
y = np.random.randint(0, 2, 1000)
# Train RandomForestClassifier
model = sklearn.ensemble.RandomForestClassifier().fit(df, y)
# Display the original class for a random test sample
x = df.iloc[0]
print(f"Factual: {x.tolist()}\nFactual class: {model.predict([x])}")from cfnow import find_tabular
# Then, we use CFNOW to generate the minimum modification to change the classification
cf_obj = find_tabular(
factual=x,
feat_types={c: 'cat' if 'cat' in c else 'cont' for c in df.columns},
has_ohe=True,
model_predict_proba=model.predict_proba,
limit_seconds=10)
# Here we can see the new class
print(f"CF: {cf_obj.cfs[0]}\nCF class: {model.predict([cf_obj.cfs[0]])}")import warnings
import pandas as pd
import numpy as np
from sklearn.preprocessing import OneHotEncoder
import sklearn.ensemble
warnings.filterwarnings("ignore", message="X does not have valid feature names, but RandomForestClassifier was fitted with feature names")
# Generate data
X = np.hstack((np.random.randint(0, 10, size=(1000, 5)), np.random.rand(1000, 3) * 100))
# One-hot encode the first 5 categorical columns
# !!!IMPORTANT!!! The naming of OHE encoding features columns MUST follow feature_value format.
# Therefore, for a feature called color with value equal to red or blue, the OHE encoding columns
# must be named color_red and color_blue. Otherwise, the CF will not be able to find the correct
# columns to modify. For binary, it is just sufficient to name refer the column as cat.
encoder = OneHotEncoder(sparse=False)
X_cat_encoded = encoder.fit_transform(X[:, :5])
names = encoder.get_feature_names_out(['cat1', 'cat2', 'cat3', 'cat4', 'cat5'])
# Combine and convert to DataFrame
df = pd.DataFrame(np.hstack((X_cat_encoded, X[:, 5:])), columns=list(names) + ['num1', 'num2', 'num3'])
# Random binary target variable
y = np.random.randint(0, 2, 1000)
# Train RandomForestClassifier
model = sklearn.ensemble.RandomForestClassifier().fit(df, y)
# Display the original class for a random test sample
x = df.iloc[0]
print(f"Factual: {x.tolist()}\nFactual class: {model.predict([x])}")from cfnow import find_tabular
# Then, we use CFNOW to generate the minimum modification to change the classification
cf_obj = find_tabular(
factual=x,
feat_types={c: 'cat' if 'cat' in c else 'cont' for c in df.columns},
has_ohe=True,
model_predict_proba=model.predict_proba,
limit_seconds=10)
# Here we can see the new class
print(f"CF: {cf_obj.cfs[0]}\nCF class: {model.predict([cf_obj.cfs[0]])}")If you use CFNOW in your research, please cite the following paper:
@article{DEOLIVEIRA2023,
title = {A model-agnostic and data-independent tabu search algorithm to generate counterfactuals for tabular, image, and text data},
journal = {European Journal of Operational Research},
year = {2023},
issn = {0377-2217},
doi = {https://doi.org/10.1016/j.ejor.2023.08.031},
url = {https://www.sciencedirect.com/science/article/pii/S0377221723006598},
author = {Raphael Mazzine Barbosa {de Oliveira} and Kenneth Sörensen and David Martens},
}