A Julia implementation of boosted trees with CPU and GPU support. Efficient histogram based algorithms with support for multiple loss functions (notably multi-target objectives such as max likelihood methods).
Input features are expected to be Matrix{Float64/Float32}. Tables/DataFrames format can be handled through MLJ (see below).
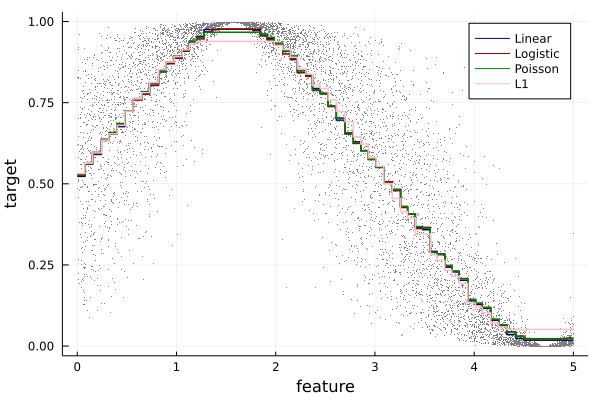
- linear
- logistic
- Poisson
- L1 (mae regression)
- Quantile
- multiclassification (softmax)
- Gaussian (max likelihood)
Set parameter device="cpu".
- linear
- logistic
- Gaussian (max likelihood)
Set parameter device="gpu".
Latest:
julia> Pkg.add("https://github.com/Evovest/EvoTrees.jl")From General Registry:
julia> Pkg.add("EvoTrees")Data consists of randomly generated float32. Training is performed on 200 iterations. Code to reproduce is here.
EvoTrees: v0.8.4 XGBoost: v1.1.1
CPU: 16 threads on AMD Threadripper 3970X GPU: NVIDIA RTX 2080
| Dimensions / Algo | XGBoost Hist | EvoTrees | EvoTrees GPU |
|---|---|---|---|
| 100K x 100 | 1.10s | 1.80s | 3.14s |
| 500K x 100 | 4.83s | 4.98s | 4.98s |
| 1M x 100 | 9.84s | 9.89s | 7.37s |
| 5M x 100 | 45.5s | 53.8s | 25.8s |
| Dimensions / Algo | XGBoost Hist | EvoTrees | EvoTrees GPU |
|---|---|---|---|
| 100K x 100 | 0.164s | 0.026s | 0.013s |
| 500K x 100 | 0.796s | 0.175s | 0.055s |
| 1M x 100 | 1.59s | 0.396s | 0.108s |
| 5M x 100 | 7.96s | 2.15s | 0.543s |
- loss: {:linear, :logistic, :poisson, :L1, :quantile, :softmax, :gaussian}
- device: {"cpu", "gpu"}
- nrounds: integer, default=10
- λ: L2 regularization, float, default=0.0
- γ: min gain for split, default=0.0
- η: learning rate, default=0.1
- max_depth: integer, default=5
- min_weight: float >= 0 default=1.0
- rowsample: float [0,1] default=1.0
- colsample: float [0,1] default=1.0
- nbins: Int, number of bins into which features will be quantilized default=64
- α: float [0,1], set the quantile or bias in L1 default=0.5
- metric: {:mse, :rmse, :mae, :logloss, :quantile, :gini, :gaussian, :none}, default=:none
- rng: random controller, either a
Random.AbstractRNGor anIntacting as a seed. Default=123.
See official project page for more info.
using StatsBase: sample
using EvoTrees
using EvoTrees: sigmoid, logit
using MLJBase
features = rand(10_000) .* 5 .- 2
X = reshape(features, (size(features)[1], 1))
Y = sin.(features) .* 0.5 .+ 0.5
Y = logit(Y) + randn(size(Y))
Y = sigmoid(Y)
y = Y
X = MLJBase.table(X)
# @load EvoTreeRegressor
# linear regression
tree_model = EvoTreeRegressor(loss=:linear, max_depth=5, η=0.05, nrounds=10)
# set machine
mach = machine(tree_model, X, y)
# partition data
train, test = partition(eachindex(y), 0.7, shuffle=true); # 70:30 split
# fit data
fit!(mach, rows=train, verbosity=1)
# continue training
mach.model.nrounds += 10
fit!(mach, rows=train, verbosity=1)
# predict on train data
pred_train = predict(mach, selectrows(X, train))
mean(abs.(pred_train - selectrows(Y, train)))
# predict on test data
pred_test = predict(mach, selectrows(X, test))
mean(abs.(pred_test - selectrows(Y, test)))Minimal example to fit a noisy sinus wave.
using EvoTrees
using EvoTrees: sigmoid, logit
# prepare a dataset
features = rand(10000) .* 20 .- 10
X = reshape(features, (size(features)[1], 1))
Y = sin.(features) .* 0.5 .+ 0.5
Y = logit(Y) + randn(size(Y))
Y = sigmoid(Y)
𝑖 = collect(1:size(X, 1))
# train-eval split
𝑖_sample = sample(𝑖, size(𝑖, 1), replace = false)
train_size = 0.8
𝑖_train = 𝑖_sample[1:floor(Int, train_size * size(𝑖, 1))]
𝑖_eval = 𝑖_sample[floor(Int, train_size * size(𝑖, 1))+1:end]
X_train, X_eval = X[𝑖_train, :], X[𝑖_eval, :]
Y_train, Y_eval = Y[𝑖_train], Y[𝑖_eval]
params1 = EvoTreeRegressor(
loss=:linear, metric=:mse,
nrounds=100, nbins = 100,
λ = 0.5, γ=0.1, η=0.1,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
pred_eval_linear = predict(model, X_eval)
# logistic / cross-entropy
params1 = EvoTreeRegressor(
loss=:logistic, metric = :logloss,
nrounds=100, nbins = 100,
λ = 0.5, γ=0.1, η=0.1,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
pred_eval_logistic = predict(model, X_eval)
# Poisson
params1 = EvoTreeCount(
loss=:poisson, metric = :poisson,
nrounds=100, nbins = 100,
λ = 0.5, γ=0.1, η=0.1,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
@time pred_eval_poisson = predict(model, X_eval)
# L1
params1 = EvoTreeRegressor(
loss=:L1, α=0.5, metric = :mae,
nrounds=100, nbins=100,
λ = 0.5, γ=0.0, η=0.1,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
pred_eval_L1 = predict(model, X_eval)# q50
params1 = EvoTreeRegressor(
loss=:quantile, α=0.5, metric = :quantile,
nrounds=200, nbins = 100,
λ = 0.1, γ=0.0, η=0.05,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
pred_train_q50 = predict(model, X_train)
# q20
params1 = EvoTreeRegressor(
loss=:quantile, α=0.2, metric = :quantile,
nrounds=200, nbins = 100,
λ = 0.1, γ=0.0, η=0.05,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
pred_train_q20 = predict(model, X_train)
# q80
params1 = EvoTreeRegressor(
loss=:quantile, α=0.8,
nrounds=200, nbins = 100,
λ = 0.1, γ=0.0, η=0.05,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0)
model = fit_evotree(params1, X_train, Y_train, X_eval = X_eval, Y_eval = Y_eval, print_every_n = 25)
pred_train_q80 = predict(model, X_train)params1 = EvoTreeGaussian(
loss=:gaussian, metric=:gaussian,
nrounds=100, nbins=100,
λ = 0.0, γ=0.0, η=0.1,
max_depth = 6, min_weight = 1.0,
rowsample=0.5, colsample=1.0, seed=123)Returns the normalized gain by feature.
features_gain = importance(model, var_names)Plot a given tree of the model:
plot(model, 2)Note that 1st tree is used to set the bias so the first real tree is #2.
EvoTrees.save(model, "data/model.bson")
model = EvoTrees.load("data/model.bson");