_ _____ _____ ____ ____ _____ ______
/ \ |_ _| |_ _| |_ || _||_ _|.' ___ |
/ _ \ | | | | | |__| | | | / .' \_|
/ ___ \ | | _ | | _ | __ | | | | |
_/ / \ \_ _| |__/ | _| |__/ | _| | | |_ _| |_\ `.___.'\
|____| |____||________||________||____||____||_____|`.____ .'
This software is currently under active development. DO NOT USE.
The easiest way to install allhic is to download the latest binary from
the releases and make sure to
chmod +x the resulting binary.
If you are using go, you can build from source with:
go get -u -t -v github.com/tanghaibao/allhic/...
go install github.com/tanghaibao/allhic/cmd/allhicPrune bamfile to remove weak links. WIP.
Extract does a fair amount of preprocessing: 1) extract inter-contig links into a more compact form, specifically into .clm; 2) extract intra-contig links and build a distribution; 3) count up the restriction sites to be used in normalization (similar to LACHESIS); 4) bundles the inter-contig links into pairs of contigs.
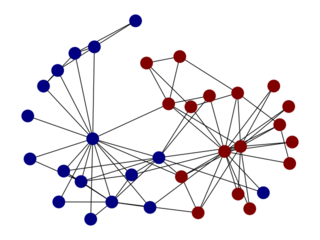
allhic extract tests/test.bam tests/test.fastaGiven a target k, number of partitions, the goal of the partitioning
is to separate all the contigs into separate clusters. As with all
clustering algorithm, there is an optimization goal here. The
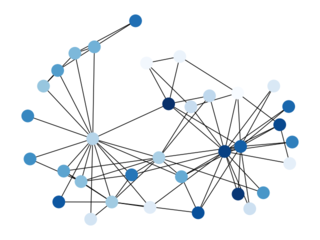
LACHESIS algorithm is a hierarchical clustering algorithm using
average links, which is the same method used by ALLHIC.
allhic partition tests/test.counts_GATC.txt tests/test.pairs.txtGiven a set of Hi-C contacts between contigs, as specified in the clmfile, reconstruct the highest scoring ordering and orientations for these contigs.
Optimize uses Genetic Algorithm (GA) to search for the best scoring solution. GA has been successfully applied to genome scaffolding tasks in the past (see ALLMAPS; Tang et al. Genome Biology, 2015).
allhic optimize tests/test.counts_GATC.g0.txt tests/test.clm
allhic optimize tests/test.counts_GATC.g1.txt tests/test.clmBuild genome release, including a .agp output and a .fasta output.
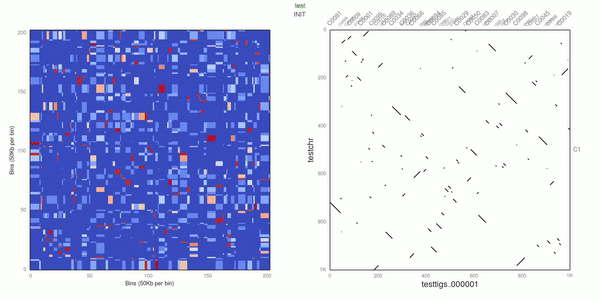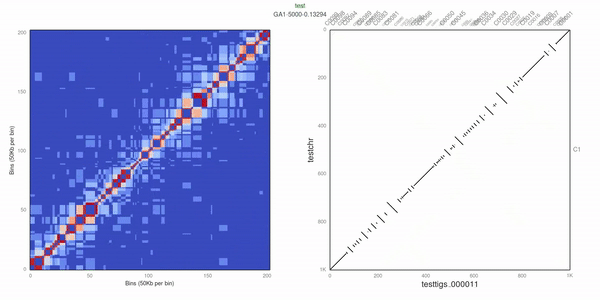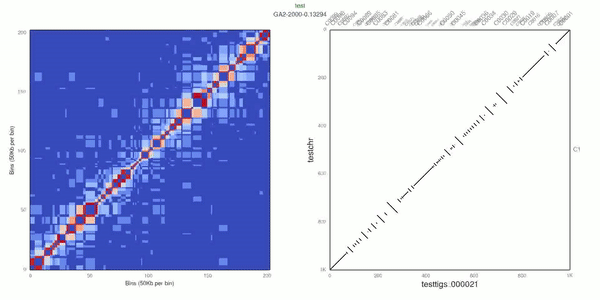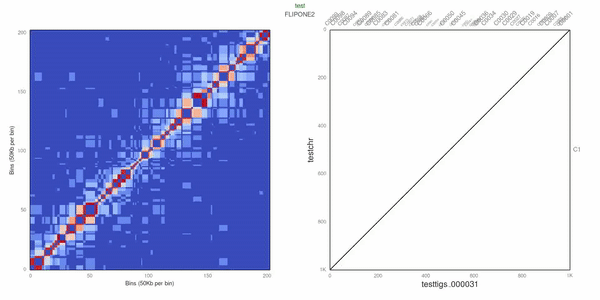
Use d3.js to visualize the heatmap.
Following the 4 steps of prune, extract, partition, optimize
allhic extract T4_Chr1/{prunning.sub.bam,seq.fasta}
allhic partition T4_Chr1/{prunning.sub.counts_GATC.txt,prunning.sub.pairs.txt} 2
allhic optimize T4_Chr1/{prunning.sub.counts_GATC.2g1.txt,prunning.sub.clm}
allhic optimize T4_Chr1/{prunning.sub.counts_GATC.2g2.txt,prunning.sub.clm}
allhic build T4_Chr/{prunning.sub.tour,seq.fasta}- Add restriction enzyme for better normalization of contig lengths
- Add partition split inside "partition"
- Use clustering when k = 1
- Isolate matrix generation to "plot"
- Add dot plot to "plot"
- Add "pipeline" to simplify execution
- Compare numerical output with Lachesis
- Improve Ler0 results
- Translate "prune" from C++ code to golang
- Add test suites