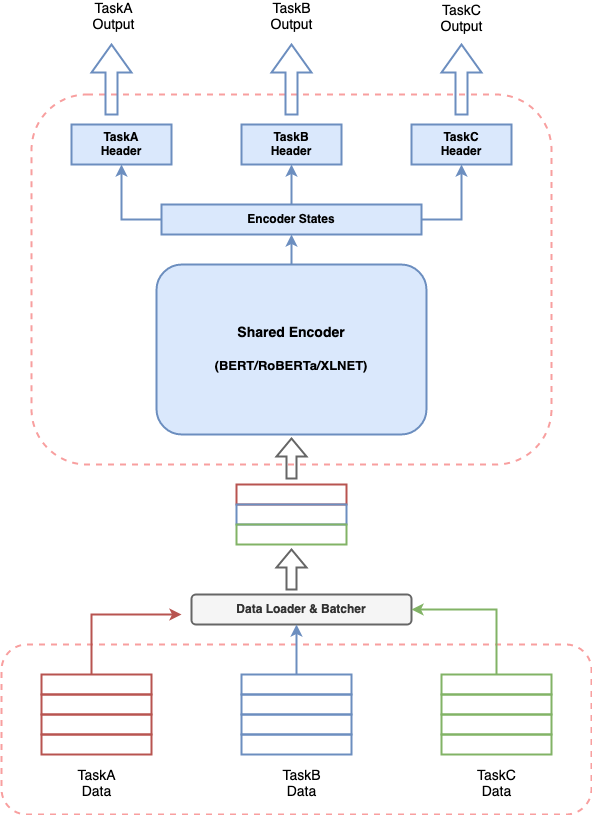
multi_task_NLP is a utility toolkit enabling NLP developers to easily train and infer a single model for multiple tasks. We support various data formats for majority of NLI tasks and multiple transformer-based encoders (eg. BERT, Distil-BERT, ALBERT, RoBERTa, XLNET etc.)
For complete documentation for this library, please refer to documentation
Any conversational AI system involves building multiple components to perform various tasks and a pipeline to stitch all components together. Provided the recent effectiveness of transformer-based models in NLP, it’s very common to build a transformer-based model to solve your use case. But having multiple such models running together for a conversational AI system can lead to expensive resource consumption, increased latencies for predictions and make the system difficult to manage. This poses a real challenge for anyone who wants to build a conversational AI system in a simplistic way.
multi_task_NLP gives you the capability to define multiple tasks together and train a single model which simultaneously learns on all defined tasks. This means one can perform multiple tasks with latency and resource consumption equivalent to a single task.
To use multi-task-NLP, you can clone the repository into the desired location on your system with the following terminal command.
>>> cd /desired/location/
>>> git clone https://github.com/hellohaptik/multi-task-NLP.git
>>> cd multi-task-NLP
>>> pip install -r requirements.txtNOTE:- The library is built and tested using Python 3.7.3. It is recommended to install the requirements in a virtual environment.
A quick guide to show how a single model can be trained for multiple NLI tasks in just 3 simple steps and with no requirement to code!!
Follow these 3 simple steps to train your multi-task model!
Task file is a YAML format file where you can add all your tasks for which you want to train a multi-task model.
TaskA: model_type: BERT config_name: bert-base-uncased dropout_prob: 0.05 label_map_or_file: -label1 -label2 -label3 metrics: - accuracy loss_type: CrossEntropyLoss task_type: SingleSenClassification file_names: - taskA_train.tsv - taskA_dev.tsv - taskA_test.tsv TaskB: model_type: BERT config_name: bert-base-uncased dropout_prob: 0.3 label_map_or_file: data/taskB_train_label_map.joblib metrics: - seq_f1 - seq_precision - seq_recall loss_type: NERLoss task_type: NER file_names: - taskB_train.tsv - taskB_dev.tsv - taskB_test.tsv
For knowing about the task file parameters to make your task file, task file parameters.
After defining the task file, run the following command to prepare the data.
>>> python data_preparation.py \
--task_file 'sample_task_file.yml' \
--data_dir 'data' \
--max_seq_len 50For knowing about the data_preparation.py script and its arguments, refer running data preparation.
Finally you can start your training using the following command.
>>> python train.py \
--data_dir 'data/bert-base-uncased_prepared_data' \
--task_file 'sample_task_file.yml' \
--out_dir 'sample_out' \
--epochs 5 \
--train_batch_size 4 \
--eval_batch_size 8 \
--grad_accumulation_steps 2 \
--log_per_updates 25 \
--save_per_updates 1000 \
--eval_while_train True \
--test_while_train True \
--max_seq_len 50 \
--silent TrueFor knowing about the train.py script and its arguments, refer running train.
Once you have a multi-task model trained on your tasks, we provide a convenient and easy way to use it for getting predictions on samples through the inference pipeline.
For running inference on samples using a trained model for say TaskA, TaskB and TaskC,
you can import InferPipeline class and load the corresponding multi-task model by making an object of this class.
>>> from infer_pipeline import inferPipeline
>>> pipe = inferPipeline(modelPath = 'sample_out_dir/multi_task_model.pt', maxSeqLen = 50)infer function can be called to get the predictions for input samples
for the mentioned tasks.
>>> samples = [ ['sample_sentence_1'], ['sample_sentence_2'] ]
>>> tasks = ['TaskA', 'TaskB']
>>> pipe.infer(samples, tasks)For knowing about the infer_pipeline, refer infer.