Read Link | AWS Cloud Deployment | Google CoLab Demo | Docker | Report Bug | Request Feature
Reporting biomarkers assessed by routine immunohistochemical (IHC) staining of tissue is broadly used in diagnostic pathology laboratories for patient care. To date, clinical reporting is predominantly qualitative or semi-quantitative. By creating a multitask deep learning framework referred to as DeepLIIF, we present a single-step solution to stain deconvolution/separation, cell segmentation, and quantitative single-cell IHC scoring. Leveraging a unique de novo dataset of co-registered IHC and multiplex immunofluorescence (mpIF) staining of the same slides, we segment and translate low-cost and prevalent IHC slides to more expensive-yet-informative mpIF images, while simultaneously providing the essential ground truth for the superimposed brightfield IHC channels. Moreover, a new nuclear-envelop stain, LAP2beta, with high (>95%) cell coverage is introduced to improve cell delineation/segmentation and protein expression quantification on IHC slides. By simultaneously translating input IHC images to clean/separated mpIF channels and performing cell segmentation/classification, we show that our model trained on clean IHC Ki67 data can generalize to more noisy and artifact-ridden images as well as other nuclear and non-nuclear markers such as CD3, CD8, BCL2, BCL6, MYC, MUM1, CD10, and TP53. We thoroughly evaluate our method on publicly available benchmark datasets as well as against pathologists' semi-quantitative scoring.
© This code is made available for non-commercial academic purposes.
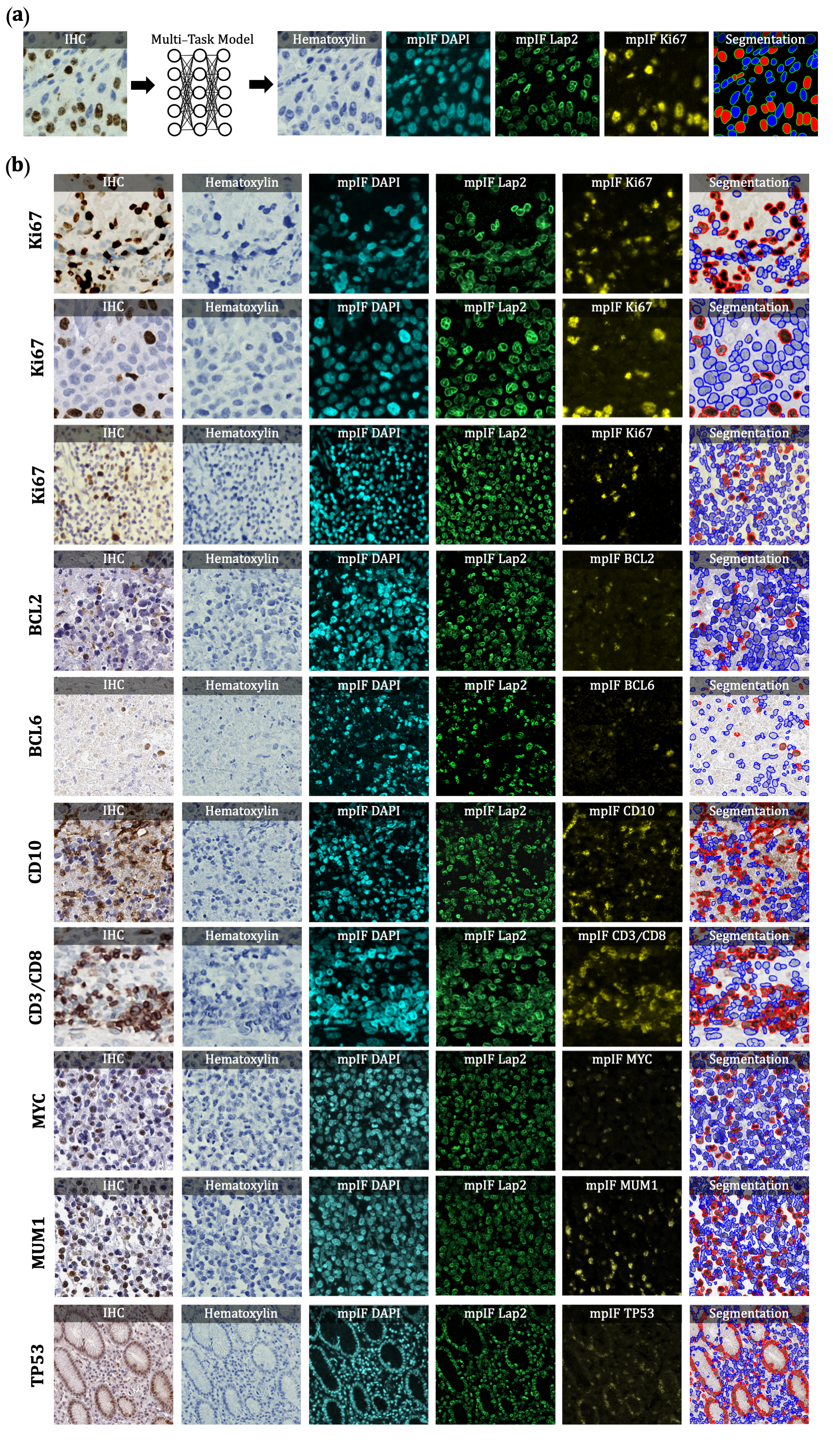 Figure 1. Overview of DeepLIIF pipeline and sample input IHCs (different
brown/DAB markers -- BCL2, BCL6, CD10, CD3/CD8, Ki67) with corresponding DeepLIIF-generated hematoxylin/mpIF modalities
and classified (positive (red) and negative (blue) cell) segmentation masks. (a) Overview of DeepLIIF. Given an IHC
input, our multitask deep learning framework simultaneously infers corresponding Hematoxylin channel, mpIF DAPI, mpIF
protein expression (Ki67, CD3, CD8, etc.), and the positive/negative protein cell segmentation, baking explainability
and interpretability into the model itself rather than relying on coarse activation/attention maps. In the segmentation
mask, the red cells denote cells with positive protein expression (brown/DAB cells in the input IHC), whereas blue cells
represent negative cells (blue cells in the input IHC). (b) Example DeepLIIF-generated hematoxylin/mpIF modalities and
segmentation masks for different IHC markers. DeepLIIF, trained on clean IHC Ki67 nuclear marker images, can generalize
to noisier as well as other IHC nuclear/cytoplasmic marker images.
Figure 1. Overview of DeepLIIF pipeline and sample input IHCs (different
brown/DAB markers -- BCL2, BCL6, CD10, CD3/CD8, Ki67) with corresponding DeepLIIF-generated hematoxylin/mpIF modalities
and classified (positive (red) and negative (blue) cell) segmentation masks. (a) Overview of DeepLIIF. Given an IHC
input, our multitask deep learning framework simultaneously infers corresponding Hematoxylin channel, mpIF DAPI, mpIF
protein expression (Ki67, CD3, CD8, etc.), and the positive/negative protein cell segmentation, baking explainability
and interpretability into the model itself rather than relying on coarse activation/attention maps. In the segmentation
mask, the red cells denote cells with positive protein expression (brown/DAB cells in the input IHC), whereas blue cells
represent negative cells (blue cells in the input IHC). (b) Example DeepLIIF-generated hematoxylin/mpIF modalities and
segmentation masks for different IHC markers. DeepLIIF, trained on clean IHC Ki67 nuclear marker images, can generalize
to noisier as well as other IHC nuclear/cytoplasmic marker images.
- Python 3.8
- Docker
DeepLIIF can be pip installed:
$ python3.8 -m venv venv
$ source venv/bin/activate
(venv) $ pip install git+https://github.com/nadeemlab/DeepLIIF.gitThe package is composed of two parts:
- a library that implements the core functions used to train and test DeepLIIF models.
- a CLI to run common batch operations including training, batch testing and Torchscipt models serialization.
You can list all available commands:
(venv) $ deepliif --help
Usage: deepliif [OPTIONS] COMMAND [ARGS]...
Options:
--help Show this message and exit.
Commands:
prepare-testing-data Preparing data for testing
prepare-training-data Preparing data for training
serialize Serialize DeepLIIF models using Torchscript
test Test trained models
train General-purpose training script for multi-task...For training, all image pairs must be 512x512 and paired together in 3072x512 images (6 images of size 512x512 stitched together horizontally). Data needs to be arranged in the following order:
XXX_Dataset
├── test
├── val
└── train
We have provided two simple functions in the CLI for preparing data for testing and training purposes.
- To prepare data for training, you need to have the paired data including IHC, Hematoxylin Channel, mpIF DAPI, mpIF
Lap2, mpIF marker, and segmentation mask in the input directory.
The script gets the address of directory containing paired data and the address of the dataset directory.
It, first, creates the train and validation directories inside the given dataset directory.
Then it reads all images in the folder and saves the pairs in the train or validation directory, based on the given
validation_ratio.
deepliif prepare-data-for-training --input_dir /path/to/input/images
--output_dir /path/to/dataset/directory
--validation_ratio 0.2
- To prepare data for testing, you only need to have IHC images in the input directory. The function gets the address of directory containing the IHC data and the address of the dataset directory. It, first, creates the test directory inside the given dataset directory. Then it reads the IHC images in the folder and saves a pair in the test directory.
deepliif prepare-data-for-testing --input_dir /path/to/input/images
--output_dir /path/to/dataset/directory
The first version of DeepLIIF model suffered from its inability to separate IHC positive cells in some large clusters, resulting from the absence of clustered positive cells in our training data. To infuse more information about the clustered positive cells into our model, we present a novel approach for the synthetic generation of IHC images using co-registered data. We design a GAN-based model that receives the Hematoxylin channel, the mpIF DAPI image, and the segmentation mask and generates the corresponding IHC image. The model converts the Hematoxylin channel to gray-scale to infer more helpful information such as the texture and discard unnecessary information such as color. The Hematoxylin image guides the network to synthesize the background of the IHC image by preserving the shape and texture of the cells and artifacts in the background. The DAPI image assists the network in identifying the location, shape, and texture of the cells to better isolate the cells from the background. The segmentation mask helps the network specify the color of cells based on the type of the cell (positive cell: a brown hue, negative: a blue hue).
In the next step, we generate synthetic IHC images with more clustered positive cells. To do so, we change the segmentation mask by choosing a percentage of random negative cells in the segmentation mask (called as Neg-to-Pos) and converting them into positive cells. Some samples of the synthesized IHC images along with the original IHC image are shown in Figure 2.
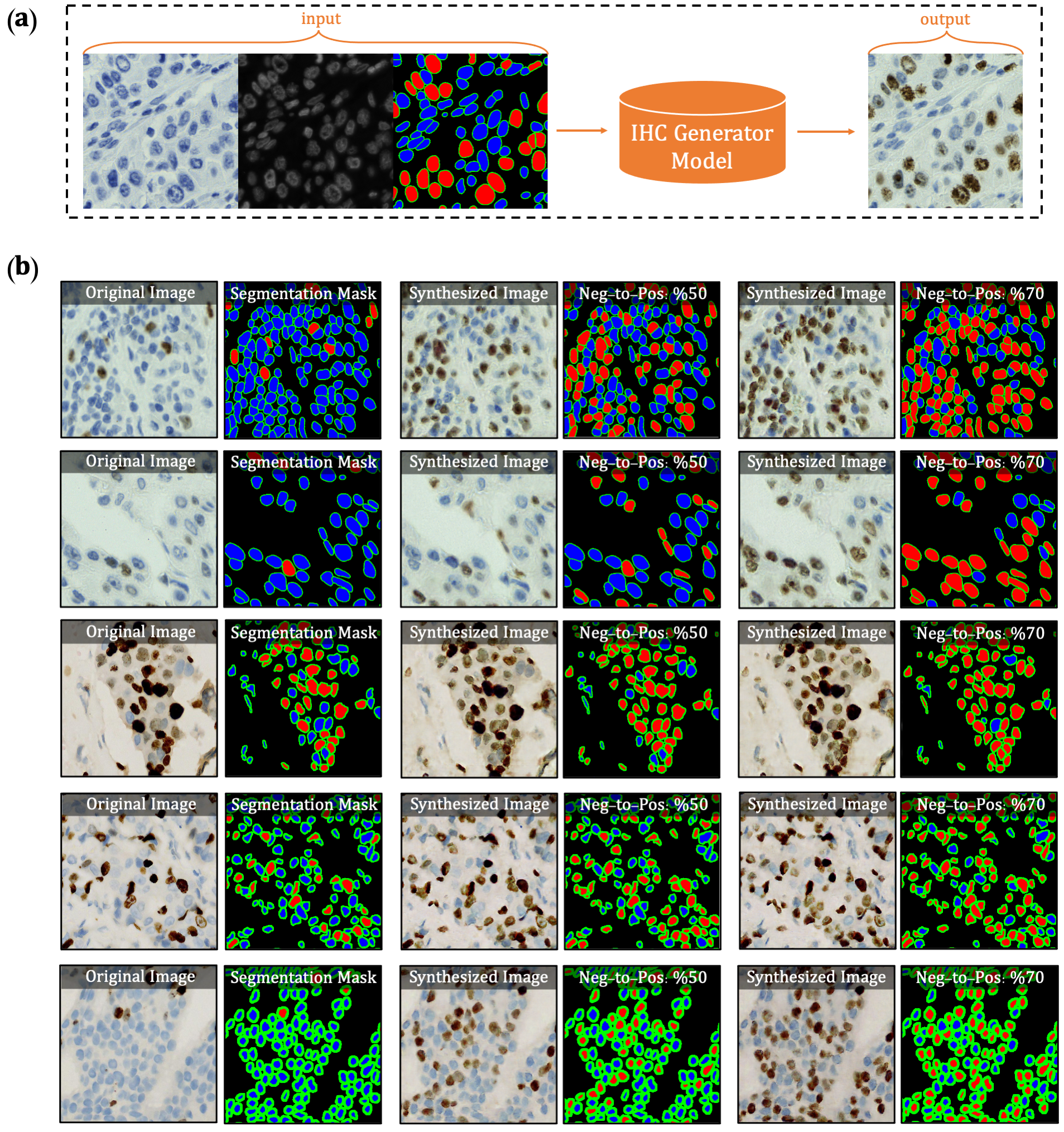 Figure 2. Overview of synthetic IHC image generation. (a) A training sample
of the IHC-generator model. (b) Some samples of synthesized IHC images using the trained IHC-Generator model. The
Neg-to-Pos shows the percentage of the negative cells in the segmentation mask converted to positive cells.
Figure 2. Overview of synthetic IHC image generation. (a) A training sample
of the IHC-generator model. (b) Some samples of synthesized IHC images using the trained IHC-Generator model. The
Neg-to-Pos shows the percentage of the negative cells in the segmentation mask converted to positive cells.
We created a new dataset using the original IHC images and synthetic IHC images. We synthesize each image in the dataset two times by setting the Neg-to-Pos parameter to %50 and %70. We re-trained our network with the new dataset. You can find the new trained model here.
To train a model:
deepliif train --dataroot /path/to/input/images
--name Model_Name
--model DeepLIIF
- To view training losses and results, open the URL http://localhost:8097. For cloud servers replace localhost with your IP.
- To epoch-wise intermediate training results, DeepLIIF/checkpoints/Model_Name/web/index.html
- Trained models will be by default save in DeepLIIF/checkpoints/Model_Name.
- Training datasets can be downloaded here.
To test the model:
deepliif test --input-dir /path/to/input/images
--output-dir /path/to/output/images
--tile-size 512
- The test results will be by default saved to DeepLIIF/results/Model_Name/test_latest/images.
- The latest version of the pretrained models can be downloaded here.
- Place the pretrained model in DeepLIIF/checkpoints/DeepLIIF_Latest_Model and set the Model_Name as DeepLIIF_Latest_Model.
- To test the model on large tissues, we have provided two scripts for pre-processing (breaking tissue into smaller tiles) and post-processing (stitching the tiles to create the corresponding inferred images to the original tissue). A brief tutorial on how to use these scripts is given.
- Testing datasets can be downloaded here.
We provide a Dockerfile that can be used to run the DeepLIIF models inside a container. First, you need to install the Docker Engine. After installing the Docker, you need to follow these steps:
- Download the pretrained model and place them in DeepLIIF/checkpoints/DeepLIIF_Latest_Model.
- Change XXX of the WORKDIR line in the DockerFile to the directory containing the DeepLIIF project.
- To create a docker image from the docker file:
docker build -t cuda/deepliif .
The image is then used as a base. You can copy and use it to run an application. The application needs an isolated environment in which to run, referred to as a container.
- To create and run a container:
docker run --gpus all -it cuda/deepliif
When you run a container from the image, the deepliif CLI will be available.
You can easily run any CLI command in the activated environment and copy the results from the docker container to the host.
If you don't have access to GPU or appropriate hardware, we have also created Google CoLab project for your convenience. Please follow the steps in the provided notebook to install the requirements and run the training and testing scripts. All the libraries and pretrained models have already been set up there. The user can directly run DeepLIIF on their images using the instructions given in the Google CoLab project.
To register the denovo stained mpIF and IHC images, you can use the registration framework in the 'Registration' directory. Please refer to the README file provided in the same directory for more details.
To train DeepLIIF, we used a dataset of lung and bladder tissues containing IHC, hematoxylin, mpIF DAPI, mpIF Lap2, and mpIF Ki67 of the same tissue scanned using ZEISS Axioscan. These images were scaled and co-registered with the fixed IHC images using affine transformations, resulting in 1667 co-registered sets of IHC and corresponding multiplex images of size 512x512. We randomly selected 709 sets for training, 358 sets for validation, and 600 sets for testing the model. We also randomly selected and segmented 41 images of size 640x640 from recently released BCDataset which contains Ki67 stained sections of breast carcinoma with Ki67+ and Ki67- cell centroid annotations (for cell detection rather than cell instance segmentation task). We split these tiles into 164 images of size 512x512; the test set varies widely in the density of tumor cells and the Ki67 index. You can find this dataset here.
We are also creating a self-configurable version of DeepLIIF which will take as input any co-registered H&E/IHC and multiplex images and produce the optimal output. If you are generating or have generated H&E/IHC and multiplex staining for the same slide (denovo staining) and would like to contribute that data for DeepLIIF, we can perform co-registration, whole-cell multiplex segmentation via ImPartial, train the DeepLIIF model and release back to the community with full credit to the contributors.
This section describes how to run DeepLIIF's inference using Torchserve workflows. Workflows con be composed by both PyTorch models and Python functions that can be connected through a DAG. For DeepLIIF there are 4 main stages (see Figure 3):
Pre-processdeserialize the image from the request and return a tensor created from it.G1-4run the ResNets to generate the Hematoxylin, DAPI, LAP2 and Ki67 masks.G51-5run the UNets and applyWeighted Averageto generate the Segmentation image.Aggregateaggregate and serialize the results and return to user.
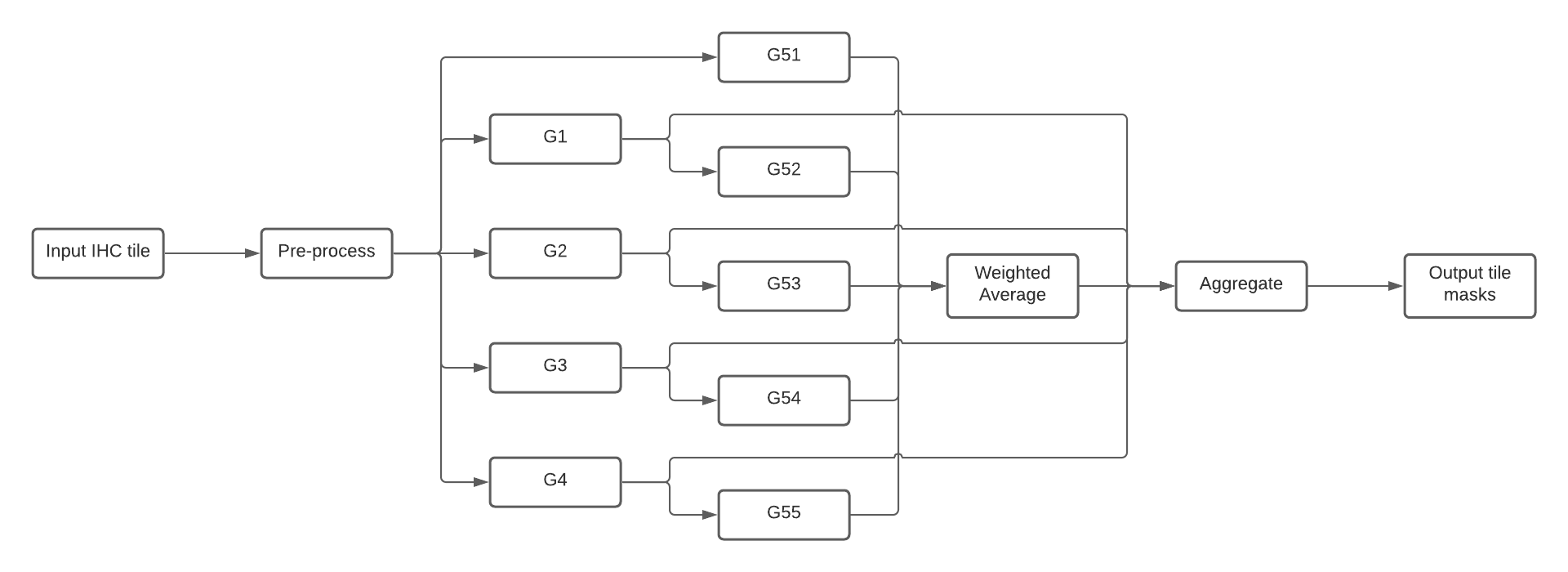 Figure 3. Composition of DeepLIIF nets into a Torchserve workflow
Figure 3. Composition of DeepLIIF nets into a Torchserve workflow
In practice, users need to call this workflow for each tile generated from the original image.
A common use case scenario would be:
- Load an IHC image and generate the tiles.
- For each tile
- Resize to 512x512 and transform to tensor.
- Serialize the tensor and use the inference API to generate all the masks
- Deserialize the results
- Stitch back the results and apply post-processing operations
The next sections show how to deploy the model server.
- Install Torchserve and torch-model-archiver following these instructions.
In MacOS, navigate to the
model-serverdirectory:
cd model-server
python3 -m venv venv
source venv/bin/activate
pip install torch torchserve torch-model-archiver torch-workflow-archiver - Download and unzip the latest version of the DeepLIIF models from zenodo.
wget https://zenodo.org/record/4751737/files/DeepLIIF_Latest_Model.zip
unzip DeepLIIF_Latest_Model.zipIn order to run the DeepLIIF nets using Torchserve, they first need to be archived as MAR files.
In this section we will create the model artifacts and archive them in the model store.
First, inside model-server create a directory to store the models.
mkdir model-storeFor every ResNet (G1, G2, G3, G4) run replacing the name of the net:
torch-model-archiver --force --model-name <Gx> \
--model-file resnet.py \
--serialized-file ./DeepLIIF_Latest_Model/latest_net_<Gx>.pth \
--export-path model-store \
--handler net_handler.py \
--requirements-file model_requirements.txtand for the UNets (G51, G52, G53, G54, G54) switch the model file from resnet.py to unet.py:
torch-model-archiver --force --model-name <G5x> \
--model-file unet.py \
--serialized-file ./DeepLIIF_Latest_Model/latest_net_<G5x>.pth \
--export-path model-store \
--handler net_handler.py \
--requirements-file model_requirements.txtOnce all the models have been packaged and made available in the model store, they can be composed into a workflow archive. Finally, create the archive for the workflow represented in Figure 3.
torch-workflow-archiver -f --workflow-name deepliif \
--spec-file deepliif_workflow.yaml \
--handler deepliif_workflow_handler.py \
--export-path model-storeOnce all artifacts are available in the model store, run the model server.
torchserve --start --ncs \
--model-store model-store \
--workflow-store model-store \
--ts-config config.propertiesAn additional step is needed to register the deepliif workflow on the server.
curl -X POST "http://127.0.0.1:8081/workflows?url=deepliif.war"The snippet below shows an example of how to cosume the Torchserve workflow API using Python.
import base64
import requests
from io import BytesIO
import torch
from deepliif.preprocessing import transform
def deserialize_tensor(bs):
return torch.load(BytesIO(base64.b64decode(bs.encode())))
def serialize_tensor(ts):
buffer = BytesIO()
torch.save(ts, buffer)
return base64.b64encode(buffer.getvalue()).decode('utf-8')
TORCHSERVE_HOST = 'http://127.0.0.1:8080'
img = load_tile()
ts = transform(img.resize((512, 512)))
res = requests.post(
f'{TORCHSERVE_HOST}/wfpredict/deepliif',
json={'img': serialize_tensor(ts)}
)
res.raise_for_status()
masks = {k: deserialize_tensor(v) for k, v in res.json().items()}Please report all issues on the public forum.
© Nadeem Lab - DeepLIIF code is distributed under Apache 2.0 with Commons Clause license, and is available for non-commercial academic purposes.
- This code is inspired by CycleGAN and pix2pix in PyTorch.
If you find our work useful in your research or if you use parts of this code, please cite our paper:
@article{ghahremani2021deepliif,
title={DeepLIIF: Deep Learning-Inferred Multiplex ImmunoFluorescence for IHC Image Quantification},
author={Ghahremani, Parmida and Li, Yanyun and Kaufman, Arie and Vanguri, Rami and Greenwald, Noah and Angelo, Michael and Hollmann, Travis J and Nadeem, Saad},
journal={bioRxiv},
year={2021},
publisher={Cold Spring Harbor Laboratory}
}
