Package vcf2circos is a python package based on Plotly which helps generating Circos plot, from a VCF file or a JSON configuration file.
See documentation and code in GitHub vcf2circos.
This package is based on PCircos code
-._ _.--'"`'--._ _.--'"`'--._ _.--'"`'--._ _
'-:`.'|`|"':-. '-:`.'|`|"':-. '-:`.'|`|"':-. '.` : '.
'. '. | | | |'. '. | | | |'. '. | | | |'. '.: '. '.
: '. '.| | | | '. '.| | | | '. '.| | | | '. '. : '. `.
' '. `.:_ | :_.' '. `.:_ | :_.' '. `.:_ | :_.' '. `.' `.
`-..,..-' `-..,..-' `-..,..-' ` `
__ ___ _
/ _|__ \ (_)
__ _____| |_ ) |___ _ _ __ ___ ___ ___
\ \ / / __| _| / // __| | '__/ __/ _ \/ __|
\ V / (__| | / /| (__| | | | (_| (_) \__ \
\_/ \___|_| |____\___|_|_| \___\___/|___/
Author: Jean-Baptiste Lamouche, Antony Le Bechec, Jin Cui
Version: 1.1
Last update: Mars 26 2023
usage: python vcf2circos.py [-h] -i INPUT -o OUTPUT [-e EXPORT] [-p OPTIONS] [-a ASSEMBLY]
optional arguments:
-h, --help show this help message and exit
-i INPUT, --input INPUT
Input vcf File
VCF SHOULD be multiallelic split to avoid trouble in vcf2circos
example: bcftools -m -any <vcf>
Format will be autodetected from file path.
Supported format:
'vcf.gz', 'vcf'
-o OUTPUT, --output OUTPUT
Output file.
Format will be autodetected from file path.
Supported format:
'png', 'jpg', 'jpeg', 'webp', 'svg', 'pdf', 'eps', 'json'
-e EXPORT, --export EXPORT
Export file.
Format is 'json'.
Generate json file from VCF input file
-p OPTIONS, --options OPTIONS
Options file in json format
-a ASSEMBLY, --assembly ASSEMBLY
Genome assembly to use for now values available (hg19, hg38, mm9, mm10)
Download package source files.
$ git clone https://github.com/bioinfo-chru-strasbourg/vcf2circos.git .
$ python -m pip install -e .
Build docker image "vcf2circos:latest"
From inside cloned repository
$ docker image build -t vcf2circos:latest .
Configuration files could be download here: vcf2circos-config (do not forget to uncompress tarball).
tar -xzf <tarballname> <folder>
Regarding where you place your configuration folder previously downloaded, you need to specify the absolute path of the Static folder in "Static" json key (which will replace default value)
If you are working with an assembly not provided in vcf2circos, you can build your own required files by following those steps:
1) Download refSeq ncbi from UCSC web server, files: ncbiRefSeqCurated.txt.gz, chromInfo.txt.gz, cytoBand.txt.gz FTP
Unzip refseq file
bgzip -d ncbirefseqfile
Sort by chromosome then position
sort -k1,1V -k2,2n file > sortedfile
A python func is available in utils.py to process ncbiRefSeqCurated.txt, run it from directory containing the python module in vcf2circos ( assembly ex: "hg19")
Create a folder with the assembly name in the Assembly folder in the config directory
python -c "from utils import formatted_refgene; formatted_refgene('path of ncbirefseq file', 'assembly')"
Creating:
genes.(assemblyname).txt
exons.(assemblyname).txt
transcripts.(assemblyname).txt
(rename each files in (type).(assemblyname).sorted.txt if it's not already done)
Decompress, sort and rename chromoInfo.txt > chr.(assemblyname).sorted.txt
Add name of columns: chr_name size
Finally cytoBand.txt.gz Add name of columns: chr_name start end band band_color Rename cytoband.txt.gz into cytoband_(assemblyname)_chr_infos.txt.gz
Be carefull for cytoband band and band_color could be inverted (band_color should contains gneg, gpos100 etc)
vcf2circos --input <vcf> --options <jsonfile> --output <outputpath>.html -a yournewassembly
$ vcf2circos --input config/Static/example.vcf.gz --options <jsonfile> --output <outputpath>.html -a <assembly hg19, hg38, mm9 or mm10>
$ docker run -it --rm vcf2circos:1.1 -i <input.vcf> -o <output.html> -p /Static/options.json -a hg19
This package allows multiple input formats:
- VCF including SNV/InDel/SV (see VCF specifications). Header needs to contain contigs (in order of appearance). See VCF example.
This package generates Circos plot in multiple formats (html, png, jpg, jpeg, webp, svg, pdf, eps, json):
- HTML file (format with customizable hover text). See HTML example
- Image files (i.e. png, jpg, jpeg, webp, svg, pdf, eps). See PNG example and PDF example
- JSON Plotly file (see Plotly documentation).
Output Circos plot sections from a VCF file:
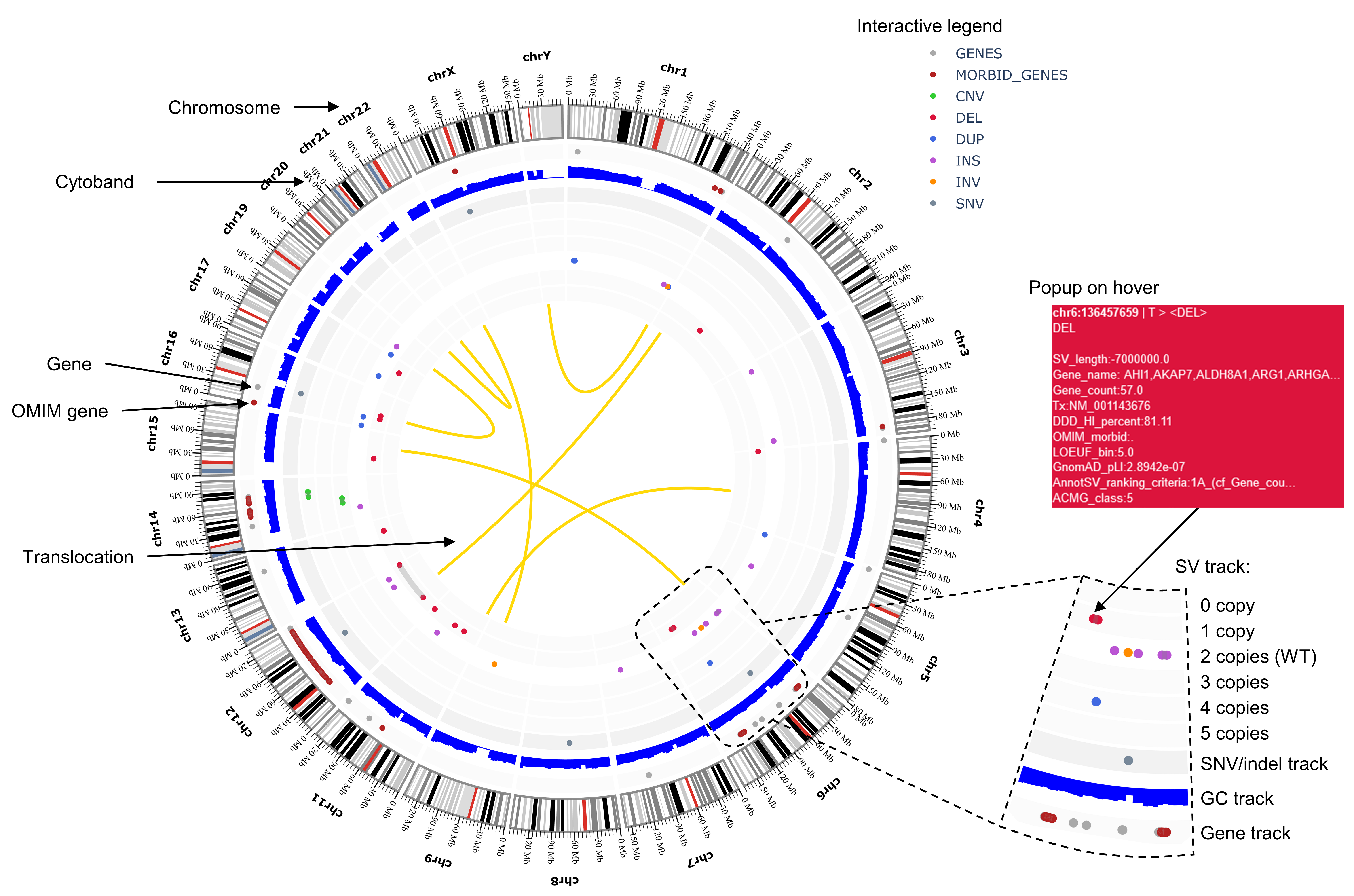
Circos plot generated from a VCF file can be configured using a JSON options file. See JSON options example.
Here is an example of a JSON options file:
{
"General": {
"title": "",
"width": 1000,
"height": 1000,
"plot_bgcolor": "white"
},
"Static": "<path of config>",
"Assembly": "hg19",
"Chromosomes": {
"list": ["chrX"],
"all": false
},
"Genes": {
"only_snv_in_sv_genes": false,
"extend": false
},
"Variants": {
"annotations": {
"fields": [
"REF",
"ALT",
"Tx",
"SV_length",
"SVLEN",
"END",
"CN",
"Gene_name",
"Gene_count",
"DDD_HI_percent",
"OMIM_morbid",
"GnomAD_pLI",
"LOEUF_bin",
"AnnotSV_ranking_criteria",
"ACMG_class"
]
},
"rings": {
"position": 0.5,
"height": 0.04,
"space": 0.01,
"nrings": 6
}
},
"Extra": [
"gc"
],
Exemple of a data tab-delimited file (STILL IN DEV): Overview of cytoband file, at terms it will be possible to add this kind of data above copy number level rings
chr_name start end band_color band
chr1 0 2300000 gneg p36.33
chr1 2300000 5400000 gpos25 p36.32
chr1 5400000 7200000 gneg p36.31
chr1 7200000 9200000 gpos25 p36.23
chr1 9200000 12700000 gneg p36.22
chr1 12700000 16200000 gpos50 p36.21
chr1 16200000 20400000 gneg p36.13
chr1 20400000 23900000 gpos25 p36.12
chr1 23900000 28000000 gneg p36.11
The "General" section is a Plotly General section, which configure main options of the Circos plot (e.g. title, size, back-ground color).
Example:
"General": {
"title": "",
"width": 1000,
"height": 1000,
"plot_bgcolor": "white"
}
The "Chromosomes" section defines information about chromosomes (e.g. contig, list of chromosomes).
Example:
"Chromosomes": {
"list": ["chr7", "chr13", "chr12", "chr14", "chr15", "chrX", "chr1", "chr17"]
}
The "list" option define the list of chromosomes to show in the Circos plot. Order of chromosome is still defined in the VCF header (in "contigs" section). If no chromosomes are listed, all chromosomes in the VCF header will be shown.
The "Genes" section defines information about Genes (e.g. refGene data, list of genes to show). These information are used to annnotate variants (SNV and SV), and are used with algorithms highlight interesting information (e.g. only SNV on CNV genes). They also can be shown in the Circos plot (below Chromosomes ring)
only_snv_in_sv_genes: display only snv indels located inside SV boundaries
extend: display genes located 1Mb in upstream and downstream of SV boundaries
"only_snv_in_sv_genes": true,
"extend": true
The "list" option defines the list of genes to show in the Circos plot, below Chromosomes/Cytoband ring. This list refers to the "gene" column in the data.
The "only_snv_in_sv_genes" option will select (and show) only SNV that are located on genes mutated with at least 1 SV.
The "Variants" section defines varaints annotations to show in each variant hover text, and positions of the varaints rings.
Example:
"Variants": {
"annotations": {
"fields": ["SVTYPE", "SVLEN"],
},
"rings": {
"position": 0.50,
"height": 0.04,
"space": 0.01,
"nrings": 6
}
}
The "annotations" option defines the annotations of variants to be shown.
The "fields" option configures the list of annotations in the hover text. If empty list if provided getting 15 first annotations in order of appearance in vcf info field. Moreover size of hover annotations is limited to 40 chars.
The "rings" option defines the "position" and "height" of SNV and SV rings, "space" between rings and the number of ring in lightgray to display.
Medical Bioinformatics Applied to Diagnosis - Strasbourg University Hospital - France