xmlBLASTparser is a lightweight PHP library for parsing an XML formatted NCBI BLAST output and rendering into a colorful HTML page. The database accession number/id in the webpage is properly hyperlinked to the external source database. Moreover, the description summary in the webpage is hyperlinked with anchor link to the corresponding alignment section. The complete list of NCBI standard sequence identifiers are tabulated below:
| Tag and Identifier Syntax | Identifier Source Description |
|---|---|
| bbm|integer | NCBI GenInfo Backbone database identifier |
| bbs|integer | NCBI GenInfo Backbone database identifier |
| dbj|coll-accession|locus | DNA Database of Japan |
| emb|coll-accession|entry | EBI EMBL Database |
| gb|coll-accession|locus | NCBI GenBank database |
| gi|integer | NCBI GenInfo Integrated Database ("jee-aye") |
| gim|integer | NCBI GenInfo Import identifier |
| gnl|database|idstring | General (user-definable) database and identifier |
| gp|coll-accession|locus_cds# | GenPept (GenBank protein) identifier |
| lcl|integer | Local (user-definable) identifier |
| oth|accession|name|release | Other (user-definable) identifier* |
| pat|country|patentid|serialno | Patent sequence identifier |
| pdb|entry|chainid | Brookhaven Protein Database |
| pir|accession|entry | Protein Information Resource International |
| prf|accession|name | Protein Research Foundation |
| ref|coll-accession|locus | NCBI RefSeq |
| sp|coll-accession|locus | SWISS-PROT database |
| tpd|coll-accession|name | Third party annotation, DDBJ |
| tpe|coll-accession|name | Third party annotation, EMBL |
| tpg|coll-accession|name | Third party annotation, GenBank |
*The NCBI has discontinued support for "oth" identifiers, but support for them is maintained in xdformat/xdget.
xmlBLASTparser can be used to parse XML file format output of the NCBI BLAST sequence alignment result obtained through any one of the following methods:
- NCBI BLAST - The XML file format output of the sequence alignment can be downloaded from the NCBI BLAST from the result page and loaded into the xmlBLASTparser PHP file. For example,
$xml = simplexml_load_file("V07E2YXG014-Alignment.xml") or die("Error: Cannot able to create object");- NCBI BLAST URL API - The XML file format content can be retrieved through PHP using NCBI BLAST URL API method. BLASTphp library is a PHP wrapper for the NCBI BLAST URL API used to stream the content of NCBI BLAST sequence alignment result into an XML file format. For example,
$out = file_get_contents("https://blast.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=Get&FORMAT_TYPE=XML&FORMAT_OBJECT=Alignment&RID=$rid");
$xml = new SimpleXMLElement($out);- Standalone NCBI BLAST - The XML file format output of the sequence alignment result can be obtained by executing the standalone NCBI BLAST executable programs such as blastn.exe, blastp.exe, blastx.exe, tblastn.exe, tblastx.exe, etc. and loaded into the xmlBLASTparser PHP file. For example,
exec('blastp.exe -db pdb -query seq.fa -remote -outfmt 5 -out out.xml');
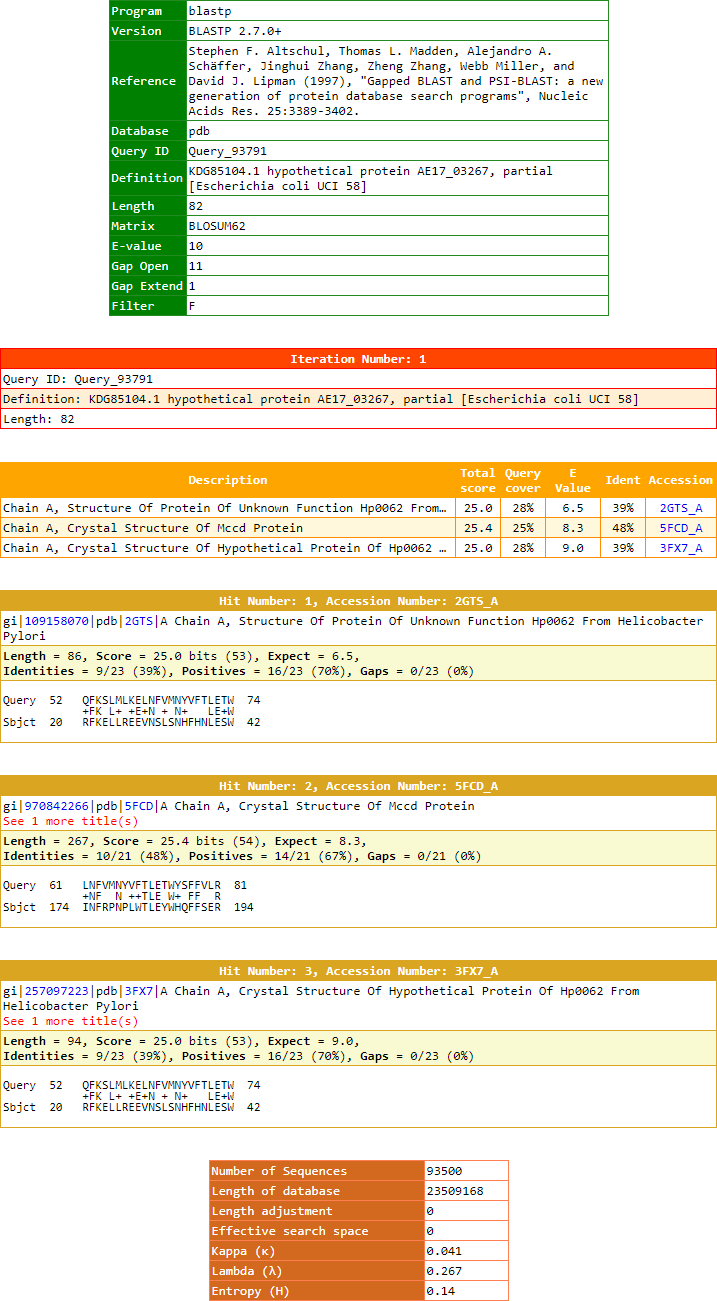
$xml = file_get_contents("out.xml");<?xml version="1.0"?>
<!DOCTYPE BlastOutput PUBLIC "-//NCBI//NCBI BlastOutput/EN" "http://www.ncbi.nlm.nih.gov/dtd/NCBI_BlastOutput.dtd">
<BlastOutput>
<BlastOutput_program>blastp</BlastOutput_program>
<BlastOutput_version>BLASTP 2.7.0+</BlastOutput_version>
<BlastOutput_reference>Stephen F. Altschul, Thomas L. Madden, Alejandro A. Sch&auml;ffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402.</BlastOutput_reference>
<BlastOutput_db>pdb</BlastOutput_db>
<BlastOutput_query-ID>Query_93791</BlastOutput_query-ID>
<BlastOutput_query-def>KDG85104.1 hypothetical protein AE17_03267, partial [Escherichia coli UCI 58]</BlastOutput_query-def>
<BlastOutput_query-len>82</BlastOutput_query-len>
<BlastOutput_param>
<Parameters>
<Parameters_matrix>BLOSUM62</Parameters_matrix>
<Parameters_expect>10</Parameters_expect>
<Parameters_gap-open>11</Parameters_gap-open>
<Parameters_gap-extend>1</Parameters_gap-extend>
<Parameters_filter>F</Parameters_filter>
</Parameters>
</BlastOutput_param>
<BlastOutput_iterations>
<Iteration>
<Iteration_iter-num>1</Iteration_iter-num>
<Iteration_query-ID>Query_93791</Iteration_query-ID>
<Iteration_query-def>KDG85104.1 hypothetical protein AE17_03267, partial [Escherichia coli UCI 58]</Iteration_query-def>
<Iteration_query-len>82</Iteration_query-len>
<Iteration_hits>
<Hit>
<Hit_num>1</Hit_num>
<Hit_id>gi|109158070|pdb|2GTS|A</Hit_id>
<Hit_def>Chain A, Structure Of Protein Of Unknown Function Hp0062 From Helicobacter Pylori</Hit_def>
<Hit_accession>2GTS_A</Hit_accession>
<Hit_len>86</Hit_len>
<Hit_hsps>
<Hsp>
<Hsp_num>1</Hsp_num>
<Hsp_bit-score>25.0238</Hsp_bit-score>
<Hsp_score>53</Hsp_score>
<Hsp_evalue>6.53601</Hsp_evalue>
<Hsp_query-from>52</Hsp_query-from>
<Hsp_query-to>74</Hsp_query-to>
<Hsp_hit-from>20</Hsp_hit-from>
<Hsp_hit-to>42</Hsp_hit-to>
<Hsp_query-frame>0</Hsp_query-frame>
<Hsp_hit-frame>0</Hsp_hit-frame>
<Hsp_identity>9</Hsp_identity>
<Hsp_positive>16</Hsp_positive>
<Hsp_gaps>0</Hsp_gaps>
<Hsp_align-len>23</Hsp_align-len>
<Hsp_qseq>QFKSLMLKELNFVMNYVFTLETW</Hsp_qseq>
<Hsp_hseq>RFKELLREEVNSLSNHFHNLESW</Hsp_hseq>
<Hsp_midline>+FK L+ +E+N + N+ LE+W</Hsp_midline>
</Hsp>
</Hit_hsps>
</Hit>
<Hit>
<Hit_num>2</Hit_num>
<Hit_id>gi|970842266|pdb|5FCD|A</Hit_id>
<Hit_def>Chain A, Crystal Structure Of Mccd Protein >gi|970842267|pdb|5FCD|B Chain B, Crystal Structure Of Mccd Protein</Hit_def>
<Hit_accession>5FCD_A</Hit_accession>
<Hit_len>267</Hit_len>
<Hit_hsps>
<Hsp>
<Hsp_num>1</Hsp_num>
<Hsp_bit-score>25.409</Hsp_bit-score>
<Hsp_score>54</Hsp_score>
<Hsp_evalue>8.26162</Hsp_evalue>
<Hsp_query-from>61</Hsp_query-from>
<Hsp_query-to>81</Hsp_query-to>
<Hsp_hit-from>174</Hsp_hit-from>
<Hsp_hit-to>194</Hsp_hit-to>
<Hsp_query-frame>0</Hsp_query-frame>
<Hsp_hit-frame>0</Hsp_hit-frame>
<Hsp_identity>10</Hsp_identity>
<Hsp_positive>14</Hsp_positive>
<Hsp_gaps>0</Hsp_gaps>
<Hsp_align-len>21</Hsp_align-len>
<Hsp_qseq>LNFVMNYVFTLETWYSFFVLR</Hsp_qseq>
<Hsp_hseq>INFRPNPLWTLEYWHQFFSER</Hsp_hseq>
<Hsp_midline>+NF N ++TLE W+ FF R</Hsp_midline>
</Hsp>
</Hit_hsps>
</Hit>
<Hit>
<Hit_num>3</Hit_num>
<Hit_id>gi|257097223|pdb|3FX7|A</Hit_id>
<Hit_def>Chain A, Crystal Structure Of Hypothetical Protein Of Hp0062 From Helicobacter Pylori >gi|257097224|pdb|3FX7|B Chain B, Crystal Structure Of Hypothetical Protein Of Hp0062 From Helicobacter Pylori</Hit_def>
<Hit_accession>3FX7_A</Hit_accession>
<Hit_len>94</Hit_len>
<Hit_hsps>
<Hsp>
<Hsp_num>1</Hsp_num>
<Hsp_bit-score>25.0238</Hsp_bit-score>
<Hsp_score>53</Hsp_score>
<Hsp_evalue>9.03233</Hsp_evalue>
<Hsp_query-from>52</Hsp_query-from>
<Hsp_query-to>74</Hsp_query-to>
<Hsp_hit-from>20</Hsp_hit-from>
<Hsp_hit-to>42</Hsp_hit-to>
<Hsp_query-frame>0</Hsp_query-frame>
<Hsp_hit-frame>0</Hsp_hit-frame>
<Hsp_identity>9</Hsp_identity>
<Hsp_positive>16</Hsp_positive>
<Hsp_gaps>0</Hsp_gaps>
<Hsp_align-len>23</Hsp_align-len>
<Hsp_qseq>QFKSLMLKELNFVMNYVFTLETW</Hsp_qseq>
<Hsp_hseq>RFKELLREEVNSLSNHFHNLESW</Hsp_hseq>
<Hsp_midline>+FK L+ +E+N + N+ LE+W</Hsp_midline>
</Hsp>
</Hit_hsps>
</Hit>
</Iteration_hits>
<Iteration_stat>
<Statistics>
<Statistics_db-num>93500</Statistics_db-num>
<Statistics_db-len>23509168</Statistics_db-len>
<Statistics_hsp-len>0</Statistics_hsp-len>
<Statistics_eff-space>0</Statistics_eff-space>
<Statistics_kappa>0.041</Statistics_kappa>
<Statistics_lambda>0.267</Statistics_lambda>
<Statistics_entropy>0.14</Statistics_entropy>
</Statistics>
</Iteration_stat>
</Iteration>
</BlastOutput_iterations>
</BlastOutput>Please feel free to sent your queries, suggestions and/or comments related to xmlBLASTparser program to [email protected] or [email protected].
xmlBLASTparser is made available under version 3 of the GNU Lesser General Public License.
Ashok Kumar, T., and Rajagopal, B. (2017). xmlBLASTparser v1.1 — a PHP based NCBI BLAST XML output parser. International Journal of Advanced Research in Computer Science. 8(8): 230-232. [Abstract] [PDF]