If you find this work helpful for your research, please consider citing:
Citation:
Liao, W., Peng, R., Hu, X., Zhang, Y., Zhou, W., Fu, X., & Lin, H. (2024). Fast forward modeling of magnetotelluric data in complex continuous media using an extended Fourier DeepONet architecture. GEOPHYSICS, 1-62. https://doi.org/10.1190/geo2023-0613.1
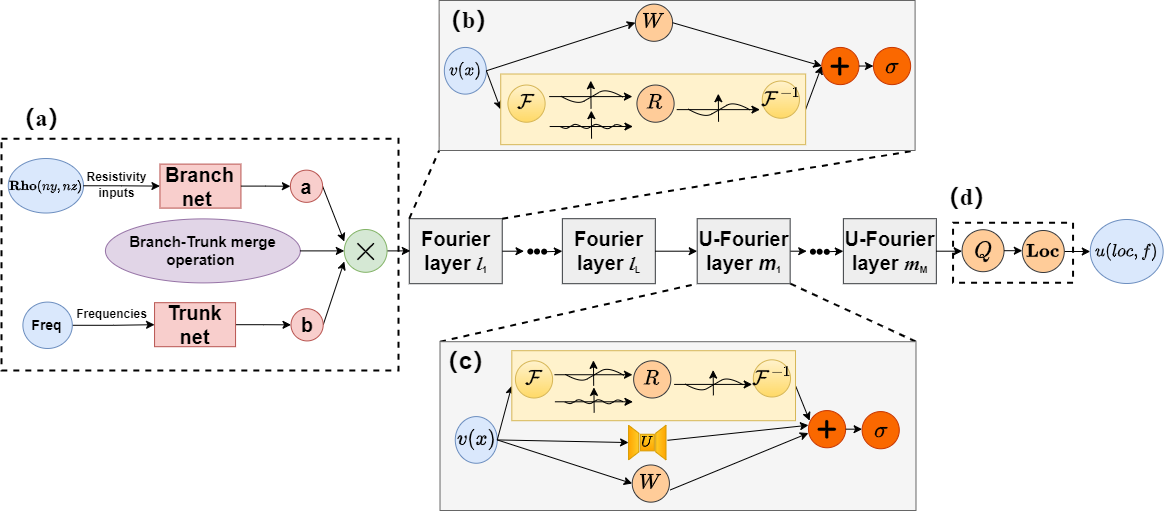 The innovative architecture of our Extended Fourier DeepONet (EFDO) neural operator network, combining the power of Fourier transforms with DeepONet for superior performance in geophysical modeling.
The innovative architecture of our Extended Fourier DeepONet (EFDO) neural operator network, combining the power of Fourier transforms with DeepONet for superior performance in geophysical modeling.
EFDO is a cutting-edge neural operator network designed specifically for geophysical modeling tasks. It combines the efficiency of Fourier transforms with the flexibility of DeepONet architecture, offering:
- 📊 Better accuracy
- 💪 Robust performance
- 🔄 Enhanced generalization
Before you dive in, make sure you have:
- 🎮 NVIDIA GPU (compute capability ≥ 6.0)
- 💾 GPU Memory: 16GB minimum (24GB recommended)
- 🗄️ System Memory: 64GB minimum (256GB recommended)
- 💽 Disk Space: 50GB minimum (512GB recommended)
You'll need these tools in your arsenal:
- 🐧 Ubuntu ≥ 18.04 LTS (or Windows ≥ 10 with WSL2)
- 🐍 Python ≥ 3.7
- 🔥 PyTorch ≥ 1.8.0
- 📚 Essential packages:
- torchinfo
- yaml
- numpy
- scipy
- pandas
- matplotlib
- jupyter notebook
💡 Pro tip: We recommend using
AnacondawithMambafor lightning-fast package installation!
First, grab Mamba from the Mambaforge download page:
bash Miniforge3-Linux-x86_64.sh -b -p ${HOME}/miniforgeAdd these magic lines to your ~/.bashrc:
# conda
if [ -f "${HOME}/miniforge/etc/profile.d/conda.sh" ]; then
source "${HOME}/miniforge/etc/profile.d/conda.sh"
fi
# mamba
if [ -f "${HOME}/miniforge/etc/profile.d/mamba.sh" ]; then
source "${HOME}/miniforge/etc/profile.d/mamba.sh"
fi
alias conda=mambaconda create -n efdo python=3.8
conda activate efdo# Install PyTorch with CUDA support
conda install pytorch torchvision torchaudio pytorch-cuda=11.7 -c pytorch -c nvidia
# Install other required packages
conda install torchinfo pyyaml numpy scipy pandas matplotlib jupyter notebook
pip install raygit clone https://github.com/CUG-EMI/EFDOQuick and parallel dataset generation:
python model_gen.py 100 50 train_gen100: Number of datasets50: Parallel processestrain_gen: Output filename
julia juliaCallGRF.jlIn this command, the generated datasets will be saved in the
datadirectory. But it is worth noting that the julia code only generates the Gaussian random field models, and you need to calculate the forward modeling results using other forward modeling codes. In this research, we use our ownMT2Djulia forward modeling code to calculate the forward modeling results. And the forward modeling code is not open source.🔧 Note: For Julia users, set up PyCall first:
ENV["PYTHON"] = "Path of the python environment"
using Pkg
Pkg.add("PyCall")
Pkg.build("PyCall")The Python forward modeling code included in this repository (which is not our original work, for more details see EFNO_GRF) has some limitations and potential issues. Therefore, we recommend:
- Use only the Gaussian random field part of this code to generate the conductivity models
- Employ other well-established forward modeling codes to calculate the MT responses
- This approach ensures more reliable and accurate dataset generation
This separation of model generation and forward computation allows for:
- Better quality control of the synthetic data
- More flexibility in choosing appropriate forward modeling algorithms
- Increased reliability of the training dataset
If you need recommendations for alternative forward modeling codes, please refer to established MT modeling software in the geophysical community.
Don't want to generate data? No problem! Download our pre-generated datasets from Google Drive datasets
- Navigate to the
codedirectory - Edit
config_EFDO.yamlwith your paths and parameters
# change to the code directory
cd code
# activate environment
conda activat efdo
# Train EFDO
python EFDO_main.py EFDO_config
# Train EFNO
python EFNO_main.py EFNO_config
# Train UFNO3d
python UFNO3d_main.py UFNO3d_configCheck out our Jupyter notebooks (*.ipynb) in the code directory for:
- Result visualization
- Performance analysis
- Model comparisons
Get a head start with our pre-trained models from Google Drive pre-trained models, you can download them and put them in the temp_model directory, and then you can use the trained models in the jupyter notebook file to predict the forward responses.
Ready to revolutionize your geophysical modeling? Let's get started! 🚀
For questions and support, open an issue in our GitHub repository or contact our team.
Welcome to the visualization guide for the EFDO Neural Network! To help researchers in geophysics better understand and utilize our open-source neural operator network, we're excited to share all the plotting scripts used in our paper.
All figures in this study were created using Generic Mapping Tools (GMT). To promote broader adoption and understanding of GMT within the research community, we provide scripts in three different formats:
- 🖥️ Traditional GMT Scripts: For command-line interface enthusiasts
- 🐍 PyGMT Scripts: For Python users seeking a more familiar syntax
- ⭐ Julia GMT Scripts: For Julia users preferring GMT functionality in Julia
💡 Pro Tip: Choose the format that matches your programming style - they all produce identical, publication-quality results!
Let's take a look at one of our key figures and how you can reproduce it using any of the three supported approaches:
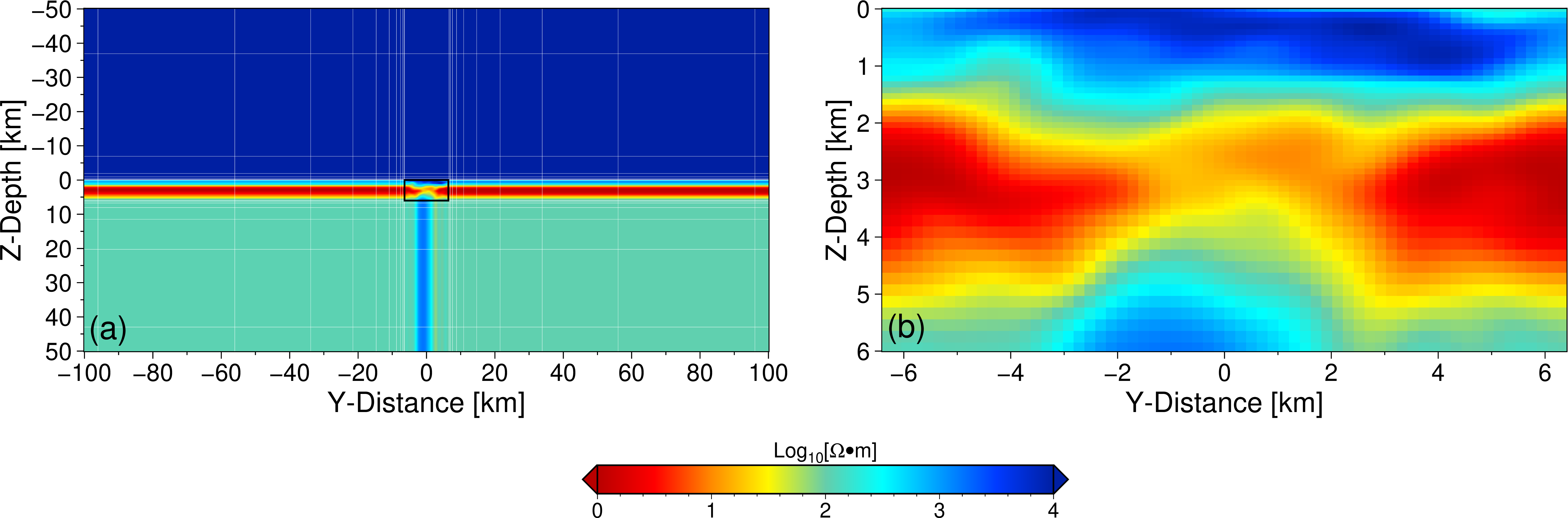 Figure: The discretization method of resistivity structures in MT forward modeling.
Figure: The discretization method of resistivity structures in MT forward modeling.
- 🖥️ GMT Script
#!/usr/bin/env bash
function preset1()
{
ymin=-100000
ymax=100000
zmin=-50000
zmax=50000
scale=1000
ymin=`echo $ymin $scale | awk '{print ($1/$2)}'`
ymax=`echo $ymax $scale | awk '{print ($1/$2)}'`
zmin=`echo $zmin $scale | awk '{print ($1/$2)}'`
zmax=`echo $zmax $scale | awk '{print ($1/$2)}'`
echo $ymin $ymax $zmin $zmax
range_yz1=$ymin/$ymax/$zmin/$zmax
}
preset1
gmt set FONT_ANNOT_PRIMARY 12p
gmt set FONT_LABEL 14p
gmt set MAP_FRAME_PEN 1p,black
# x=0 slice
gmt begin Figure5 pdf,png
gmt makecpt -Cthermal.cpt -T0/4.0/0.01 -Z -H > rbow.cpt
gmt grdconvert wholeRangeGrids.grd out.grd
gmt grdedit out.grd -R$range_yz1 -Gresult.nc
gmt basemap -Bxa20f10+l"Y-Distance [km]" -Bya10f5+l"Z-Depth [km]" -BWStr -R$range_yz1 -JX12c/-6c
gmt grdimage -R$range_yz1 -JX12c/-6c result.nc -Crbow.cpt
# y grid lines
gmt plot -Wfaint,white line_y.txt -Frs
# z grid lines
gmt plot -Wfaint,white line_z.txt -Frs
# the core region
gmt plot -W0.8p <<EOF
-6.4 0
-6.4 6.0
6.4 6.0
6.4 0
-6.4 0
EOF
gmt text -F+f16p,Helvetica << EOF
-93 43 (a)
EOF
function preset2()
{
ymin=-6400
ymax=6400
zmin=0
zmax=6000
scale=1000
ymin=`echo $ymin $scale | awk '{print ($1/$2)}'`
ymax=`echo $ymax $scale | awk '{print ($1/$2)}'`
zmin=`echo $zmin $scale | awk '{print ($1/$2)}'`
zmax=`echo $zmax $scale | awk '{print ($1/$2)}'`
echo $ymin $ymax $zmin $zmax
range_yz2=$ymin/$ymax/$zmin/$zmax
}
preset2
gmt grdconvert coreRangeGrids.grd out.grd
gmt grdedit out.grd -R$range_yz2 -Gresult.nc
gmt basemap -Bxa2f1+l"Y-Distance [km]" -Bya1f1+l"Z-Depth [km]" -BWStr -R$range_yz2 -JX12c/-6c -X14
gmt grdimage -R$range_yz2 -JX12c/-6c result.nc -Crbow.cpt
gmt text -F+f16p,Helvetica << EOF
-5.95 5.55 (b)
EOF
gmt colorbar -R$range_yz1 -Crbow.cpt -DjTC+w8c/0.5c+o-7.0c/8.0c+ml+e -N -Bxaf+l"Log@-10@-[@~W\267@~m]" -By -FONT_ANNOT_PRIMARY=15p
rm out.grd rbow.cpt *.nc
gmt end- 🐍 PyGMT Script
import pygmt
def preset1():
# Convert range units to kilometers
ymin, ymax, zmin, zmax = -100, 100, -50, 50 # Assuming these values are already in kilometers
return ymin, ymax, zmin, zmax
def preset2():
# Convert range units to kilometers
ymin, ymax, zmin, zmax = -6.4, 6.4, 0, 6 # Again, assuming these values are already in kilometers
return ymin, ymax, zmin, zmax
# Set file and directory paths
dir_path = "./"
cpt_file_in = f"{dir_path}thermal.cpt"
cpt_file_out = f"{dir_path}rbow.cpt"
result1_grid = f"{dir_path}result1.grd"
result2_grid = f"{dir_path}result2.grd"
out_pdf_png = f"{dir_path}Figure5_py"
# Set range by calling functions
ymin, ymax, zmin, zmax = preset1()
range_yz1 = f"{ymin}/{ymax}/{zmin}/{zmax}"
pygmt.config(FONT_ANNOT_PRIMARY="12p", FONT_LABEL="14p", MAP_FRAME_PEN="1p,black")
fig = pygmt.Figure()
pygmt.makecpt(cmap=cpt_file_in, series="0/4.0/0.01", continuous=True, output=cpt_file_out)
# Convert grid range using grdedit in original GMT, currently, PyGMT does not support grdedit and grdconvert modules
# gmt grdedit wholeRangeGrids.grd -R-100/100/-50/50 -Gresult1.grd
fig.grdimage(grid=result1_grid, region=range_yz1, projection="X12c/-6c", cmap=cpt_file_out, frame=["xa20f10+lY-Distance [km]", "ya10f10+lDepth [km]", "WStr"])
fig.plot(data=f"{dir_path}line_y.txt", pen="faint,white")
fig.plot(data=f"{dir_path}line_z.txt", pen="faint,white")
fig.plot(data=[[-6.4, 0], [-6.4, 6.0], [6.4, 6.0], [6.4, 0], [-6.4, 0]], pen="0.8p")
fig.text(x=-93, y=43, text="(a)", font="16p,Helvetica")
ymin, ymax, zmin, zmax = preset2()
range_yz2 = f"{ymin}/{ymax}/{zmin}/{zmax}"
# Convert grid range using grdedit in original GMT, currently, PyGMT does not support grdedit and grdconvert modules
# gmt grdedit coreRangeGrids.grd -R-6.4/6.4/0/6 -Gresult2.grd
fig.shift_origin(xshift="14c")
fig.grdimage(grid=result2_grid, region=range_yz2, projection="X12c/-6c", cmap=cpt_file_out, frame=["xa2f1+lY-Distance [km]", "ya1f1+lDepth [km]", "WStr"])
fig.text(x=-5.95, y=5.55, text="(b)", font="16p,Helvetica")
pygmt.config(FONT_ANNOT_PRIMARY="14p", FONT_LABEL="14p")
fig.colorbar(cmap=cpt_file_out, frame=["xaf+lLog@-10@-[@~W\267@~m]", "y"], position="jTC+w8c/0.5c+o-7.0c/8.0c+ml+e")
# Save in PDF and PNG formats
fig.savefig(f"{out_pdf_png}.pdf")
fig.savefig(f"{out_pdf_png}.png")- ⭐ Julia GMT Script
using GMT
function preset1()
ymin = -100000
ymax = 100000
zmin = -50000
zmax = 50000
scale = 1000
ymin /= scale
ymax /= scale
zmin /= scale
zmax /= scale
println("$ymin $ymax $zmin $zmax")
range_yz1 = string(ymin, "/", ymax, "/", zmin, "/", zmax)
return range_yz1
end
function preset2()
ymin = -6400
ymax = 6400
zmin = 0
zmax = 6000
scale = 1000
ymin /= scale
ymax /= scale
zmin /= scale
zmax /= scale
println("$ymin $ymax $zmin $zmax")
range_yz2 = string(ymin, "/", ymax, "/", zmin, "/", zmax)
return range_yz2
end
range_yz1 = preset1()
range_yz2 = preset2()
gmtbegin("Figure5_jl", fmt="pdf,png")
# Create a color palette
C = makecpt("-Cthermal.cpt -T0/4/0.1 -Z > rbow.cpt")
# Figure (a)
data1 = grdedit("wholeRangeGrids.grd", region=range_yz1)
basemap!(region=range_yz1, figsize=(12, -6), frame=(axes=:WStr,),
xaxis=(annot=20, ticks=10, label=:"Y-Distance [km]"),
yaxis=(annot=10, ticks=5, label=:"Z-Depth [km]"),
par=(FONT_ANNOT_PRIMARY=12, FONT_LABEL=14, MAP_FRAME_PEN="1p,black",)
)
grdimage!(data1, region=range_yz1, figsize=(12, -6), cmap="rbow.cpt")
plot!("line_y.txt", pen=(0.1, :White))
plot!("line_z.txt", pen=(0.1, :White))
boxline = [-6.4 0;-6.4 6.0;6.4 6.0;6.4 0;-6.4 0]
plot!(boxline, pen="0.8p", close=true)
text!(["(a)"], font=16, x=-93, y=43)
# Figure (b)
data2 = grdedit("coreRangeGrids.grd", region=range_yz2)
basemap!(region=range_yz2, figsize=(12, -6), frame=(axes=:WStr,),
xaxis=(annot=2, ticks=1, label=:"Y-Distance [km]"),
yaxis=(annot=1, ticks=1, label=:"Z-Depth [km]"),
par=(FONT_ANNOT_PRIMARY=12, FONT_LABEL=14,MAP_FRAME_PEN="1p,black",),
xshift=14
)
grdimage!(data2, region=range_yz2, figsize=(12, -6), cmap="rbow.cpt")
text!(["(b)"], font=16, x=-5.95, y=5.55)
# Adding the colorbar
gmtset(FONT_ANNOT_PRIMARY = "14p,Helvetica,black", FONT_LABEL = "14p,black")
colorbar!(xaxis=(annot=1, ticks=0.2, label=:"Log@-10@-[@~W\267@~m]"),
pos=(paper=true, anchor=(-5,-2.5), size=(8,0.5), horizontal=true,
move_annot=:l, triangles=true))
gmtend()As you can see, each approach achieves the same result while catering to different programming preferences and workflows.
We recommend using conda to create dedicated environments:
# Setting up GMT
conda create -n gmt python=3.12
conda activate gmt
conda install -c conda-forge gmt
# if `gmt --version` displays some error, please try the following way
conda remove gdal sqlite
conda install -c conda-forge gmt gdal sqlite
conda update gmt
# Setting up PyGMT
conda create -n pygmt python=3.12
conda activate pygmt
conda install numpy scipy pandas xarray netcdf4 packaging pygmtFor Julia users, it's just one line:
using Pkg
Pkg.add("GMT") # That's all!💡 Recommendation: We suggest using the latest version of GMT in WSL2 with Ubuntu for the best experience!
All figure scripts are located in the Plotting_scripts directory:
cd Plotting_scripts/Figure5
# For GMT users
conda activate gmt
bash Figure5.sh
# For Python users
conda activate pygmt
python Figure5.py
# For Julia users
julia Figure5.jlWe also provide Geophysics_plotting_scripts.ipynb which includes:
- 📈 Code for all figures
- 🔍 Detailed step-by-step explanations
- 📝 Interactive environment
- 📚 Visit the GMT official documentation
- 💬 Join GMT community discussions
- 🐛 Submit issues on GitHub