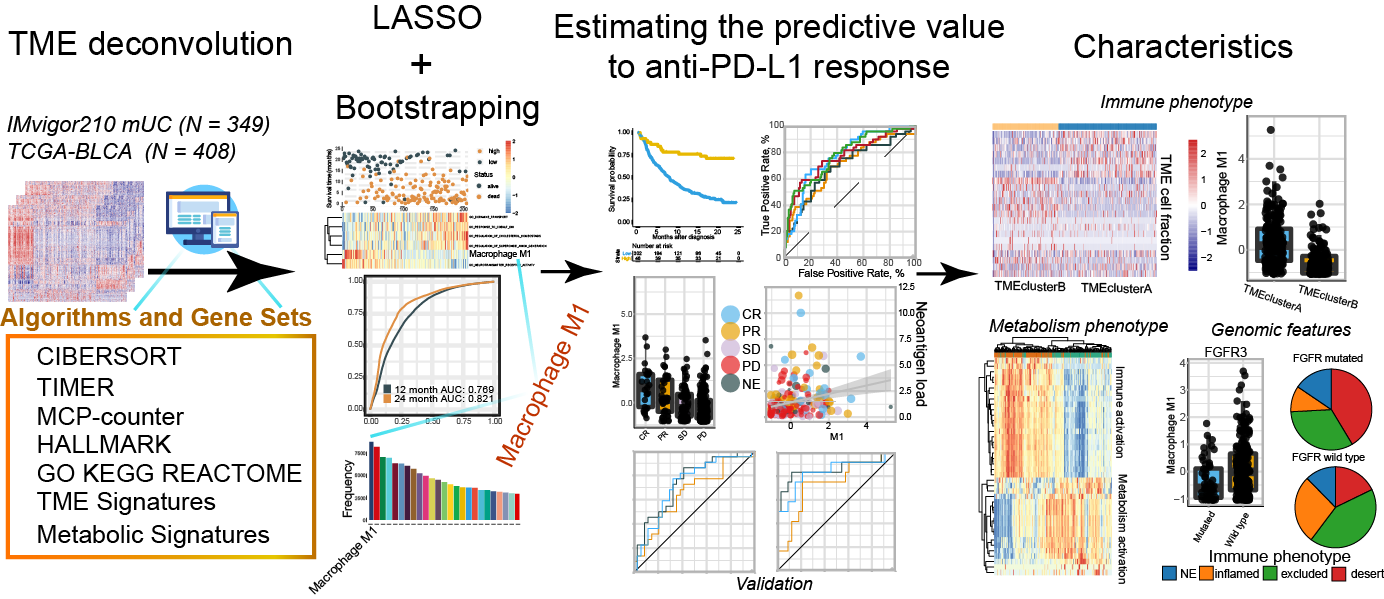
Blasso: Integrating LASSO regression and bootstrapping algorithm to find best prognostic or predictive feature
The package is not yet on CRAN. You can install from Github:
if (!requireNamespace("devtools", quietly = TRUE))
install.packages("devtools")
if (!requireNamespace("Blasso", quietly = TRUE))
devtools::install_github("DongqiangZeng0808/Blasso")Loading packages and main function in the package:
library(Blasso)
#> 载入需要的程辑包:glmnet
#> 载入需要的程辑包:Matrix
#> Loaded glmnet 4.1-2
#> 载入需要的程辑包:ggplot2
#> 载入需要的程辑包:tibble
#> 载入需要的程辑包:survival
#> 载入需要的程辑包:RColorBrewer
#> 载入需要的程辑包:progress
#> 载入需要的程辑包:stringr
#> ==========================================================================
#> Blasso v0.1.0 For help: https://github.com/DongqiangZeng0808/Blasso
#>
#> If you use Blasso in published research, please cite:
#> Macrophage correlates with immunophenotype and predicts anti-PD-L1
#> response of urothelial cancer. Theranostics 2020; 10(15):7002-7014.
#> ==========================================================================
help("best_predictor_cox")
#> starting httpd help server ...
#> done
help("best_predictor_binomial")Supplementary data
data("target")
head(target)
#> ID status time
#> 1 SAM00b9e5c52da9 1 1.9055441
#> 2 SAM0257bbbbd388 1 15.6386037
#> 3 SAM025b45c27e05 1 8.7720739
#> 4 SAM032c642382a7 1 2.4969199
#> 5 SAM04c589eb3fb3 0 0.6899384
#> 6 SAM0571f17f4045 1 4.5338809
data("features")
features[1:5,1:5]
#> ID Glycosphosphatidylinositol_PCA Macrophage_M1_cibersort
#> 1 SAM00b9e5c52da9 -0.3791156 -0.9651426
#> 2 SAM0257bbbbd388 1.3471887 -0.8690076
#> 3 SAM025b45c27e05 -0.1356366 -0.9915367
#> 4 SAM032c642382a7 -1.5168052 0.8050212
#> 5 SAM04c589eb3fb3 -3.0750685 0.6753930
#> GO_CATECHOLAMINE_TRANSPORT GO_DOPAMINE_TRANSPORT
#> 1 -0.05571328 -0.2575771
#> 2 -0.27535773 -0.3974832
#> 3 0.74345430 0.5631046
#> 4 -1.69420060 -1.4923073
#> 5 -1.58320438 -1.3433413res<-best_predictor_cox(target_data = target,
features = features,
status = "status",
time = "time",
nfolds = 10,
permutation = 300,
show_progress = FALSE)head(res$res, n = 10)
#> res1 Freq
#> 1 Macrophage M1 cibersort 270
#> 2 GO RESPONSE TO COBALT ION 239
#> 3 GO REGULATION OF CHOLESTEROL HOMEOSTASIS 217
#> 4 GO NEUROTRANSMITTER RECEPTOR ACTIVITY 211
#> 5 Glycosphosphatidylinositol PCA 194
#> 6 GO IMIDAZOLE CONTAINING COMPOUND METABOLIC PROCESS 187
#> 7 Dendritic cell activated cibersort 185
#> 8 GO CELL CELL ADHESION VIA PLASMA MEMBRANE ADHESION MOLECULES 178
#> 9 GO REGULATION OF DEFENSE RESPONSE TO BACTERIUM 153
#> 10 T cell CD4 posi memory activated cibersort 144res<-best_predictor_binomial(target_data = target,
features = features,
response = "status",
nfolds = 10,
permutation = 300,
show_progress = FALSE)head(res$res, n = 10)
#> res1 Freq
#> 1 Macrophage M1 cibersort 285
#> 2 GO RESPONSE TO COBALT ION 273
#> 3 GO REGULATION OF CHOLESTEROL HOMEOSTASIS 245
#> 4 GO SOMATIC STEM CELL POPULATION MAINTENANCE 240
#> 5 GO NEUROTRANSMITTER RECEPTOR ACTIVITY 231
#> 6 GO M BAND 225
#> 7 Dendritic cell activated cibersort 215
#> 8 GO RECEPTOR SIGNALING PROTEIN SERINE THREONINE KINASE ACTIVITY 212
#> 9 GO CENTROSOME LOCALIZATION 211
#> 10 Glycosphosphatidylinositol PCA 200sessionInfo()
#> R version 4.1.1 (2021-08-10)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 19041)
#>
#> Matrix products: default
#>
#> locale:
#> [1] LC_COLLATE=Chinese (Simplified)_China.936
#> [2] LC_CTYPE=Chinese (Simplified)_China.936
#> [3] LC_MONETARY=Chinese (Simplified)_China.936
#> [4] LC_NUMERIC=C
#> [5] LC_TIME=Chinese (Simplified)_China.936
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] Blasso_0.1.0 stringr_1.4.0 progress_1.2.2 RColorBrewer_1.1-2
#> [5] survival_3.2-11 tibble_3.1.4 ggplot2_3.3.5 glmnet_4.1-2
#> [9] Matrix_1.3-4
#>
#> loaded via a namespace (and not attached):
#> [1] lattice_0.20-44 prettyunits_1.1.1 ps_1.6.0 assertthat_0.2.1
#> [5] rprojroot_2.0.2 digest_0.6.27 foreach_1.5.1 utf8_1.2.2
#> [9] R6_2.5.1 evaluate_0.14 highr_0.9 pillar_1.6.2
#> [13] rlang_0.4.11 rstudioapi_0.13 callr_3.7.0 rmarkdown_2.11
#> [17] labeling_0.4.2 desc_1.3.0 devtools_2.4.2 splines_4.1.1
#> [21] munsell_0.5.0 compiler_4.1.1 xfun_0.25 pkgconfig_2.0.3
#> [25] pkgbuild_1.2.0 shape_1.4.6 htmltools_0.5.2 tidyselect_1.1.1
#> [29] codetools_0.2-18 fansi_0.5.0 crayon_1.4.1 dplyr_1.0.7
#> [33] withr_2.4.2 grid_4.1.1 gtable_0.3.0 lifecycle_1.0.0
#> [37] DBI_1.1.1 magrittr_2.0.1 scales_1.1.1 cli_3.0.1
#> [41] stringi_1.7.4 cachem_1.0.6 farver_2.1.0 fs_1.5.0
#> [45] remotes_2.4.0 testthat_3.0.4 ellipsis_0.3.2 vctrs_0.3.8
#> [49] generics_0.1.0 iterators_1.0.13 tools_4.1.1 glue_1.4.2
#> [53] purrr_0.3.4 hms_1.1.0 processx_3.5.2 pkgload_1.2.2
#> [57] fastmap_1.1.0 yaml_2.2.1 colorspace_2.0-2 sessioninfo_1.1.1
#> [61] memoise_2.0.0 knitr_1.34 usethis_2.0.1Zeng D, Ye Z, Wu J, Zhou R, Fan X, Wang G, Huang Y, Wu J, Sun H, Wang M, Bin J, Liao Y, Li N, Shi M, Liao W. Macrophage correlates with immunophenotype and predicts anti-PD-L1 response of urothelial cancer. Theranostics 2020; 10(15):7002-7014. doi:10.7150/thno.46176
Contact: E-mail any questions to [email protected]