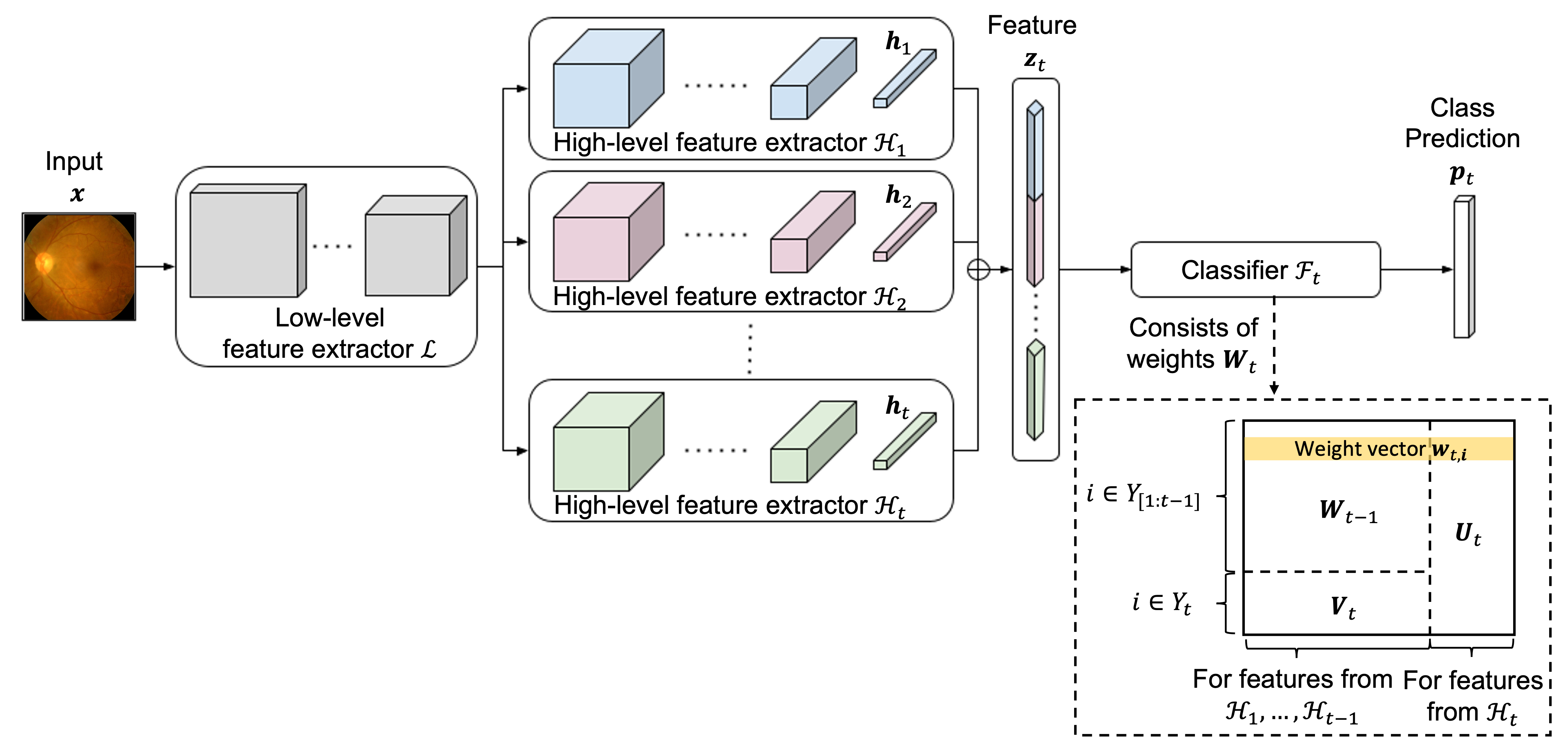
This repository is the official implementation of Leveraging Old Knowledge to Continually Learn New Classes in Medical Images.
This implementation has been tested on Python 3.8, Ubuntu 20.04, CUDA 11.4.
To install package requirements:
pip install -r requirements.txt
To prepare datasets, please refer to the respective folders for instructions:
- CCH5000: data/colorectal
- HAM10000: data/ham10000
- EyePACS: data/diabetic
For implementation details of our proposed method, please refer to inclearn/models/der.py. It is a generalized framework which uses dynamically expandable representations.
To run experiments on CCH5000 with three different class orders and the protocol of 1 classes per step using our approach:
python3 -minclearn --options options/der/derE3_colorectal.yaml options/data/colorectal_3orders.yaml \
--initial-increment 4 --increment 1 --fixed-memory --workers 4 --batch-size 32 \
--device <gpu_id> --label derE3_colorectal_4steps1 \
--data-path data/colorectalLikewise, for HAM10000 with three different class orders and the protocol of 1 class per step:
python3 -minclearn --options options/der/derE3_ham10000.yaml options/data/ham10000_3orders.yaml \
--initial-increment 3 --increment 1 --fixed-memory --workers 4 --batch-size 32 \
--device <gpu_id> --label derE0_ham10000_3steps1 \
--data-path data/ham10000And, for EyePACS:
python3 -minclearn --options options/der/derE3_diabetic.yaml options/data/diabetic_3orders.yaml \
--initial-increment 3 --increment 1 --fixed-memory --workers 4 --batch-size 128 \
--device <gpu_id> --label derE3_diabetic_3steps1 \
--data-path data/diabeticTo conduct experiments using the baseline methods, you just need to replace the first argument of --options with the path to the corresponding hyperparameter file, which can be found under the folder options.
By default, no checkpoints will be saved. To save them after training of each task, add the argument --save-model task.
Your training and evaluation results can be found under the folder results/dev, including the summary of the tasks in each run.
For reproduction of the results without training from scratch, we included the outputs from our training under the folder results/pretrain. To generate the images and results found in the paper, follow the steps under the notebook metric.ipynb and gradcam.ipynb.
Below shows a summary of our model performance:
| Dataset | New classes per step | Average incremental accuracy |
|---|---|---|
| CCH5000 | 1 | 94.5 |
| CCH5000 | 2 | 94.6 |
| HAM10000 | 1 | 78.1 |
| HAM10000 | 2 | 82.0 |
| EyePACS | 1 | 81.9 |
Thanks for the great code base by Arthur Douillard and Shipeng Yan.