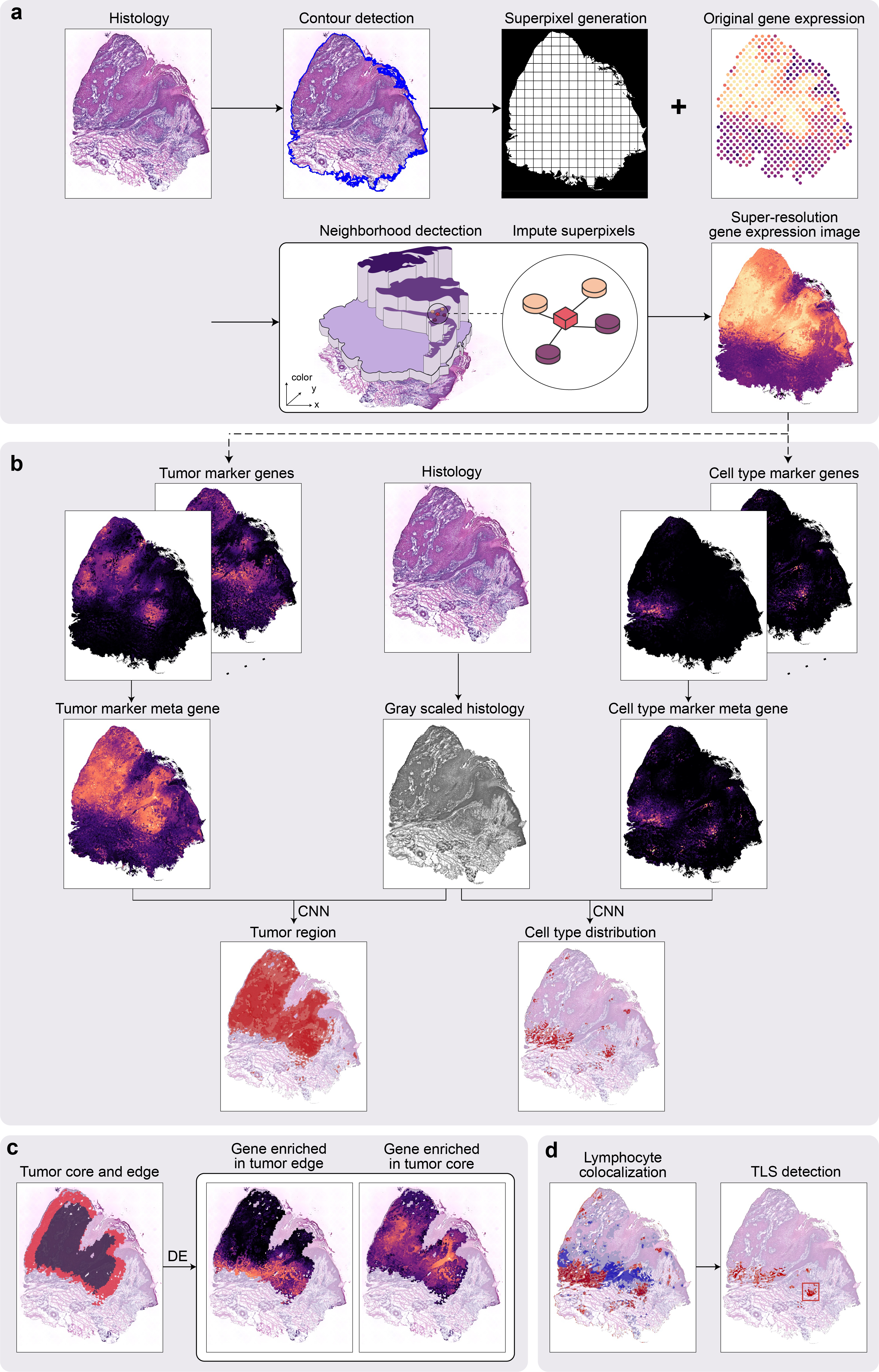
TESLA (Tumor Edge Structure and Lymphocyte multi-level Annotation) is a machine learning framework for multi-level tissue annotation on the histology image with pixel-level resolution in Spatial Transcriptomics (ST). By integrating information from high-resolution histology image, TESLA can impute gene expression at superpixels and fill in missing gene expression in tissue gaps. The increased gene expression resolution makes it possible to treat gene expression data as images, which enabled the integration with histological features for joint tissue segmentation and annotation of different cell types directly on the histology image with pixel-level resolution. Additionally, TESLA can detect unique structures of tumor immune microenvironment such as Tertiary Lymphoid Structures (TLSs), , separate a tumor into core and edge to examine their cellular compositions, expression features, and molecular processes. TESLA has been evaluated on five cancer datasets. Our results consistently showed that TESLA can generate high-quality super-resolution gene expression images, which facilitated the downstream multi-level tissue annotation.
TESLA has been used for cancer genomics data analysis in numerous top-tier journals.
The TESLA package is an implementation of multi-level tissue annotation on the histology image with pixel-level resolution in spatial transcriptomics. With TESLA, you can:
- Enhance the gene expression resolution to pixel resolution the same as the histology image;
- Annotate tumor region at pixel resolution;
- Annotate user-defined cell types at pixel resolution;
- Characterize the intra-tumor heterogeneity, e.g, the tumor leading edge, tumor core and edge, tumor subtypes.
For tutorial, please refer to: https://github.com/jianhuupenn/TESLA/blob/main/tutorial/tutorial.md
A Jupyter Notebook of the tutorial is accessible from : https://github.com/jianhuupenn/TESLA/blob/main/tutorial/tutorial.ipynb
Please install jupyter in order to open this notebook.
Toy data and results can be downloaded at: https://drive.google.com/drive/folders/1hC6ldkxmZX0yiCWZR57iMXjWIIm9qUJU?usp=sharing
Python support packages: torch, pandas, numpy, scipy, scanpy > 1.5, anndata, sklearn, cv2.
Environment 1:
- System: Mac OS 10.13.6
- Python: 3.7.0
- Python packages: pandas = 1.1.3, numpy = 1.18.1, torch=1.5.1,louvain=0.6.1,scipy = 1.4.1, scanpy = 1.5.1, anndata = 0.6.22.post1, sklearn = 0.22.1, cv2=4.5.1.
Environment 2:
- System: Anaconda
- Python: 3.7.9
- Python packages: pandas = 1.1.3, numpy = 1.20.2, python-igraph=0.8.3, torch=1.6.0,louvain=0.7.0, scipy = 1.5.2, scanpy = 1.6.0, anndata = 0.7.4, sklearn = 0.23.3, cv2=4.5.1
Souce code: Github
We are continuing adding new features. Bug reports or feature requests are welcome.
Last update: 11/20/2021, version 1.0.0
Please consider citing the following reference: