BiBERTa
Deep learning-assisted to accelerate the discovery of donor/acceptor pairs for high-performance organic solar cells
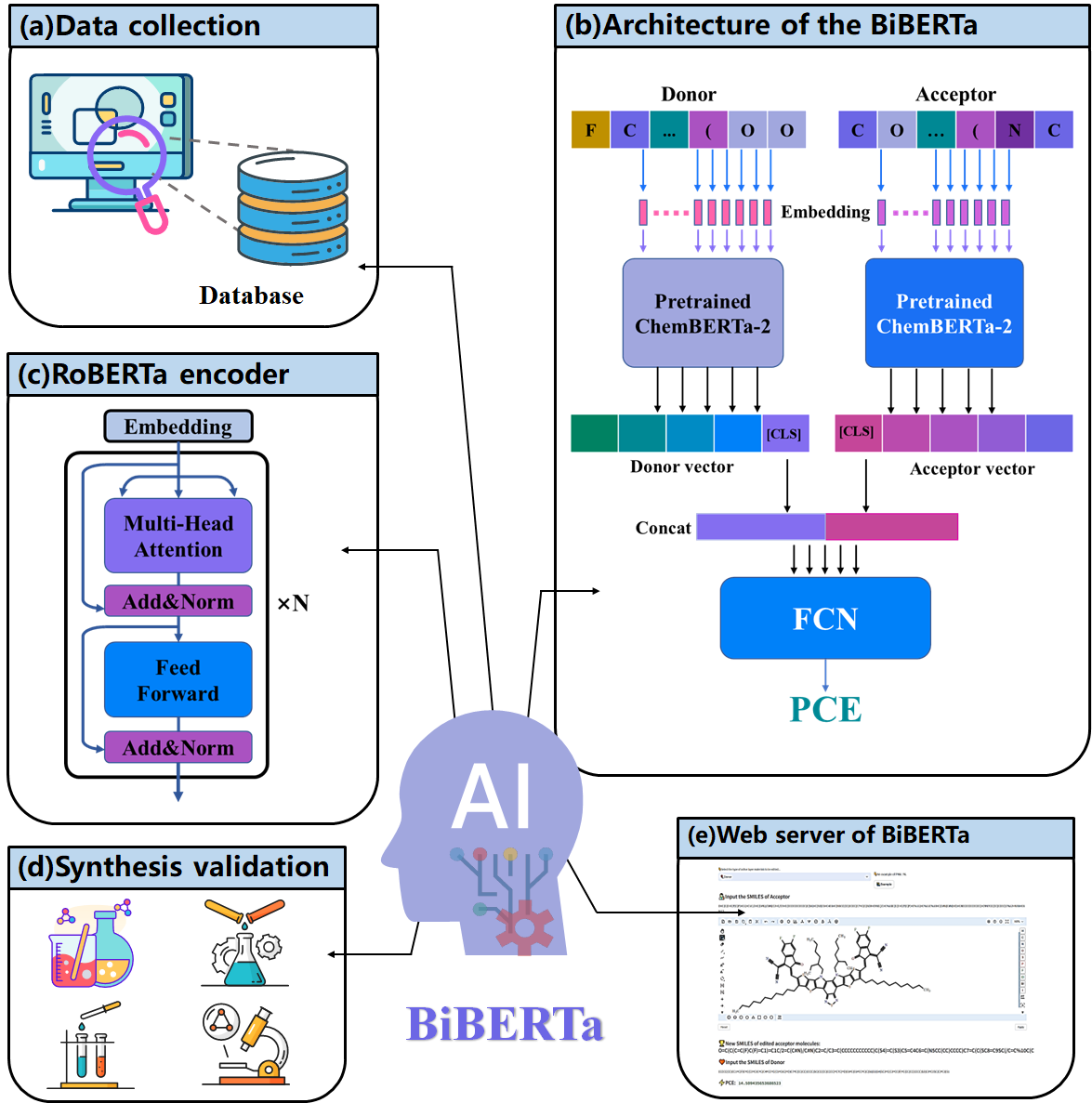
It is a deep learning-based framework built for new donor/acceptor pairs discovery. The framework contains data collection section, PCE prediction section and molecular discovery section. Specifically, a large D/A pair dataset was built by collecting experimental data from literature. Then, a novel RoBERTa-based bi-encoder model (BiBERTa) was developed for PCE prediction by using the SMILES of donor and acceptor pairs as the input. Two pretrained ChemBERTa2 encoders were loaded as initial parameters of the bi-encoder. The model was trained, tested and validated on the experimental dataset.
We recommend to use conda and pip.
By using the requirements.txt file, it will install all the required packages.
git clone --depth=1 https://github.com/JinYSun/biberta.git
cd biberta
conda create --name biberta python=3.8
conda activate biberta
conda install pip
pip install -r requirements.txt
-- train: contains the codes for training the model.
-- predict: contain the code for screening large-scale DAPs.
-- test.ipynb: contain the tutorials on predicting with models.
-- run: contain the code to predict the performance of DAP one by one.
-- dataset/OSC: contain the dataset for training/testing/validating the model.
-- Demo/example.ipynb: contain the code to show how to train/test/screen the model.
cd BiBERTa
python train.py
large scale screening by inputting a file
python -c "import screen; screen.smiles_aas_test(r'BiBERTa/dataset/OSC/test.csv')"
predicting by input the SMILES of donor and acceptor
python -c "import run; run.smiles_adp_test ('CCCCC(CC)CC1=C(F)C=C(C2=C3C=C(C4=CC=C(C5=C6C(=O)C7=C(CC(CC)CCCC)SC(CC(CC)CCCC)=C7C(=O)C6=C(C6=CC=C(C)S6)S5)S4)SC3=C(C3=CC(F)=C(CC(CC)CCCC)S3)C3=C2SC(C)=C3)S1','CCCCC(CC)CC1=CC=C(C2=C3C=C(C)SC3=C(C3=CC=C(CC(CC)CCCC)S3)C3=C2SC(C2=CC4=C(C5=CC(Cl)=C(CC(CC)CCCC)S5)C5=C(C=C(C)S5)C(C5=CC(Cl)=C(CC(CC)CCCC)S5)=C4S2)=C3)S1') "
test.ipynb: contain the tutorials to show how to use the model for prediction.
The example.ipynb was used to show the whole process of abcBERT. The files in Demo were used to test that the codes work well. The parameters (such as epochs, dataset size) were set to small numbers to show how the abcBERT worked.
The Discussion folder contains the scripts for evaluating the PCE prediction performance. We compared sevaral methods widely used in molecular property prediction.
Jinyu Sun. E-mail: [email protected]