Chandrasekhar Anisotropic Rotating Plummer.
This repository allows for the computation of the diffusion coefficients and the diffusion rates of a rotating, anisotropic Plummer cluster using the orbit-averaged Chandrasekhar theory.
The parametrization of velocity anisotropy follows Dejonghe (1987), and is described by a parameter
The parametrization rotation follows the Lynden-Bell demon from Lynden-Bell (1960), and described by
where
This repository makes use of the Julia Language (see Bezanson et al. (2017)), whose repository is located at JuliaLang.
A GPU-accelerated version of this code, making use of the CUDA.jl library, is available at the repository CARP_GPU.
Install Julia by following the instructions at julialang.org/downloads/.
To invoke Julia in the Terminal, you need to make sure that the julia command-line program is in your PATH. See here for detailed instructions.
Open the terminal in the folder packages and type
$ julia Install-pkg.jl
to install the following packages:
ArgParseHypergeometricFunctionsSpecialFunctionsStaticArraysInterpolationsHDF5PlotsLaTeXStrings
DO NOT INTERRUPT THE DOWNLOADING OF THE PACKAGES !!!!
To compute a mapping of the diffusion rate code/compute/. These script will compute a 2D map in action space and store it in a .hf5 file in the folder code/data.
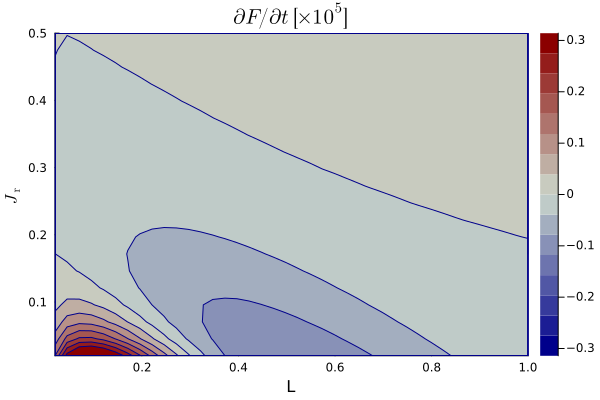
As an example, Compute_dFdt_JrL.jl will compute a 2D map in
This script is parallelized. The full list of arguments can be found in the file code/sources/julia/Args.jl.
Here is an example of a console command to launch the computation in
$ cd ./code/compute
$ julia -t 12 Compute_dFdt_JrL.jl --q 0.0 --a 0.1
The resulting file will be created in the folder code/data under the name
Dump_dFdt_JrL_Map_q_0.0_a_0.1.hf5 in about 30 seconds for these parameters.
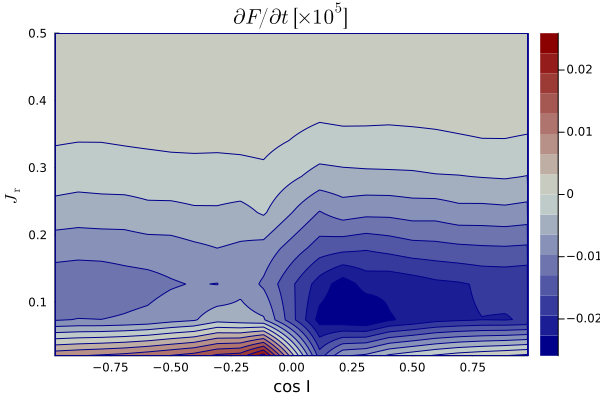
One may also be interested in samlping the
$ cd ./code/compute
$ julia -t 12 Compute_dFdt_JrCosI.jl --q 0.0 --a 0.1 --nbw 20 --nbphi 200 --nbvartheta 50
The resulting file will be created in the folder code/data under the name
Dump_dFdt_JrCosI_Map_q_0.0_a_0.1.hf5 in about 120 seconds for these parameters.
After having sampled a 2D-slice of the action space, one may plot the data using the Julia scripts located at code/compute/.
As an example, PlotContour_JrL.jl will compute a 2D map in
$ cd ./code/compute
$ julia PlotContour_JrL.jl --q 0.0 --a 0.1
The resulting file will be created in the folder code/graphs under the name
Map_dFdt_JrL_q_0.0_a_0.1.png.
Similarly, PlotContour_JrCosI.jl will compute a 2D map in
$ cd ./code/compute
$ julia PlotContour_JrCosI.jl --q 0.0 --a 0.1
The resulting file will be created in the folder code/graphs under the name
Map_dFdt_JrCosI_q_0.0_a_0.1.png.