By examining the results across a diverse set of searches, our developed JustRetrieval a methodology to systematically probe the the Best Match algorithm for preferentially bias. We found publications with white first and last authors and lower author counts were preferred. We also found that publications that were are government funded, those likely to be cited in clinical trials, and those highly influential within their field were more likely to be promoted. Finally, we found that Best Match searches contained significantly more overlap within their references sections than a control search.
The goal of JustRetrieval is to describe any potential biases that exist in search results based on PubMed Best Search Algorithm in comparing the retrieved results between different pages as well as to search results from a different search algorithm (publication date sort algorithm). We achieved this by selecting various correlates that can be broadly divided into author attributes and publication attributes. PubMed and iCite APIs were queried to retrieve top 20 results for these two algorithms and the results were compared across the select correlated.
Understanding and mitigating bias in medical information is critical to promoting health equity in biomedical research and practice. This includes how scholarly research is retrieved and presented to the end users. PubMed is a free search engine used by millions of users around the world to access ever expanding scholarly literature and resources in biomedical sciences. The NCBI/NLM at the NIH develops and maintains PubMed including the search and retrieval aspects to provide the most relevant information to user searches. In 2018, NCBI team has implemented new search algorithm called Best Match [1] to provide more relevant search results replacing an algorithm based on date sort order. The Best Match algorithm accomplishes this by combining user intelligence (past user searches with relevance ranking factors) with machine learning techniques. As with any AI/ML algorithm, it is important to ensure that the search results are fair and unbiased to avoid or mitigate bisinformation or biased information [2]. Some of the biases in algorithms may stem from the use of past feedback that can lead to "rich-get-richer dynamics", i.e. items ranked higher can reinforce feedback loops influencing future ranking [3]. With Best Match algorithm, any biases in training data, for instance with user searches and clicks, can by default manifest as an inequitous, common view of the world presented to diverse users [4].
The focus for this project is to answer the following research questions:
- RQ1: Is there a correlation between various author attributes and retrieved best match search results?
- RQ2: Is there a difference between search results between best search and date order search by author attributes?
- RQ3: Is there a correlation between various publication attributes and retrieved best match search results?
- RQ4: Is there a difference between search results between best search vs. date order search by Publication Attributes?
The author attributes that were considered are: gender, race, institutional affiliation, country of origin, and author authority (e.g., research impact based on number of hits). The publication attributes that were considered are: NIH funding, language of publication, reading level, diversity of references, associated data, number of authors and affiliations, clinical translation, research focus (human, animal or mol/cellular), and covid-focus.
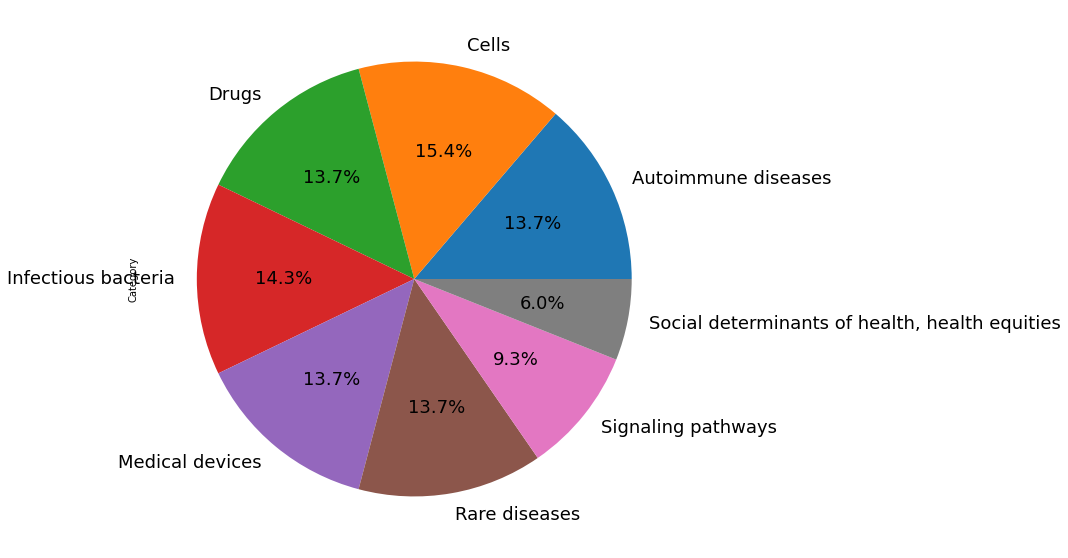
We filtered out certain publication types such as books, errata, and commentary and have relied on both past user search behaviors as well as custom search keywords across these categories: Rare diseases, signaling pathways, social determinants of health and health equities, list of autoimmune diseases, list of cells, infectious bacteria, list of medical devices, and list of drugs.
- Read the CSV files of search terms to use as search parameters for PubMed API.
- Connect to the APIs (PubMed's eUtils - both BestMatch and Publication Date Sort endpoints) to retrieve PMIDs and corresponding data.
- Query and retrieve (both Best Match and Date Sort implementations) author and publication attributes for 1st 2 pages (1st 20 results).
3.1 For author attributes, use additional packages, Python's Ethnicolr and Genderize, to derive gender and race.
3.2 For publication attributes, use content from iCite for additional data points. - With the help of Pandas and other data management libraries, write feature outputs for rollups of (query, first / second page and Pubmed ranking algorithms) to feature files.
- Use Python's statistics libraries (i.e., Pinguoin, Scipy) to:
5.1 Compare the author and publication attributes in retrieved results for the first page (1st 10 results) with second page (2nd 10 results) of PubMed Best Match algorithm results.
5.2 Compare the author and publication attributes in retrieved results (just 1st page) between the PubMed Best Search and PubMed Date Sort algorithms. - Display the results using Python's visualization libraries (i.e., Seaborn, Matplotlib) and write observations.
- Manually curated list of search terms and filters (CSV file)
- PubMed API
- iCite API
This is the data pipeline that describes the datasets used to produce our analysis. Specific data files are described in detail below.
graph LR;
team1_search_strats_search_terms.csv-->search.py
search.py-->pmids.csv
pmids.csv-->fetch_articles_from_pmids.ipynb
fetch_articles_from_pmids.ipynb-->pmid_xmls
pmids.csv-->fetch_article_data.py
fetch_article_data.py-->pmid_data.csv
pmid_data.csv-->feature_english_lang.py
feature_english_lang.py-->is_english_only.csv
pmid_data.csv-->feature_rcr.ipynb
feature_rcr.ipynb-->RCR.csv
pmid_data.csv-->journal_country.ipynb
journal_country.ipynb-->country_journal.csv
pmid_data.csv-->race_ethnicity_name.ipynb
race_ethnicity_name.ipynb-->RaceEthGender.csv
pmid_data.csv-->feature_apt_biomedicine.ipynb
feature_apt_biomedicine.ipynb-->apt.csv
pmid_data.csv-->build_feature_affiliation_count.py
build_feature_affiliation_count.py-->affiliation_count.csv
pmid_data.csv-->build_feature_author_count.py
build_feature_author_count.py-->author_count.csv
pmid_data.csv-->build_feature_reading_level.py
build_feature_reading_level.py-->readability_fk_score_abstract.csv
build_feature_reading_level.py-->readability_fk_score_title_abstract_combined.csv
build_feature_reading_level.py-->readability_fk_score_title.csv
pmid_data.csv-->data_visualizations_bl.ipynb
apt.csv-->data_visualizations_bl.ipynb
feature_rcr.ipynb-->human_animal_molcellular.csv
data_visualizations_bl.ipynb-->apt_score.png
country_journal.csv-->data_visualizations_bl.ipynb
data_visualizations_bl.ipynb-->journal_country_of_origin.png
is_english_only.csv-->data_visualizations_bl.ipynb
data_visualizations_bl.ipynb-->is_english_only.png
human_animal_molcellular.csv-->data_visualizations_bl.ipynb
readability_fk_score_abstract.csv-->data_visualizations_bl.ipynb
data_visualizations_bl.ipynb-->readability_abstract.png
readability_fk_score_title.csv-->data_visualizations_bl.ipynb
data_visualizations_bl.ipynb-->readability_title.png
pmid_data.csv-->feature_reference_diversity.ipynb
feature_reference_diversity.ipynb-->reference_diversity.csv
data_visualizations_bl.ipynb-->first_author_gender.png
data_visualizations_bl.ipynb-->first_author_race.png
data_visualizations_bl.ipynb-->last_author_gender.png
data_visualizations_bl.ipynb-->last_author_race.png
data_visualizations_bl.ipynb-->reference_diversity.png
data_visualizations_bl.ipynb-->relative_citation_ratio.png
pmid_data.csv-->extract_funding.py
extract_funding.py-->funding_and_covid_data.tsv
pmid_data.csv-->features.py
features.py-->author_affiliation.csv
features.py-->author_country.csv
pmid_data.csv-->data_visualizations_bl.ipynb
RaceEthGender.csv-->data_visualizations_bl.ipynb
reference_diversity.csv-->data_visualizations_bl.ipynb
RCR.csv-->data_visualizations_bl.ipynb
author_count.csv-->data_visualizations_bl.ipynb
data_visualizations_bl.ipynb-->author_count.png
affiliation_count.csv-->data_visualizations_bl.ipynb
data_visualizations_bl.ipynb-->affiliation_count.png
funding_and_covid_data.tsv-->data_visualizations_bl.ipynb
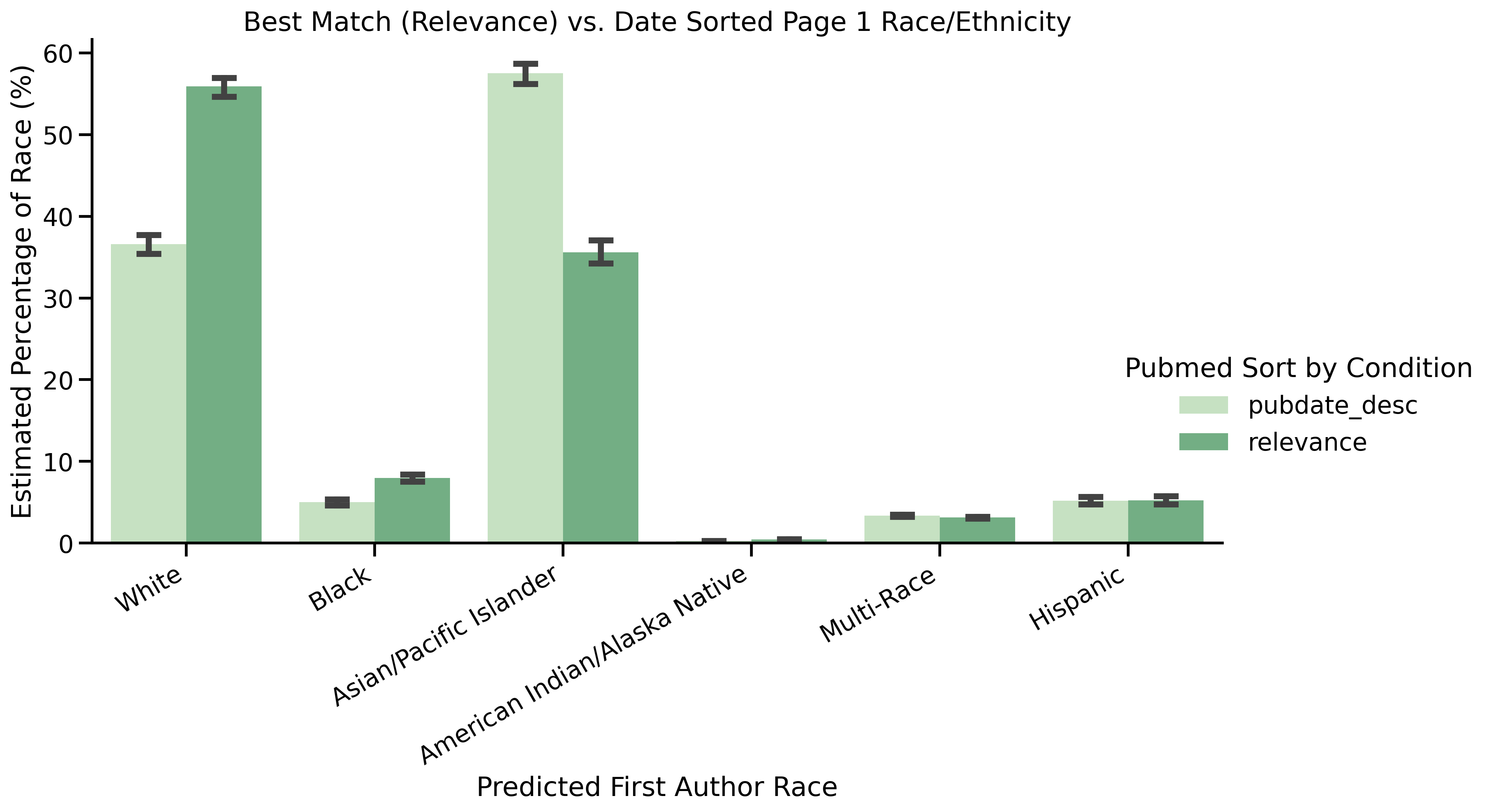
data_visualizations_bl.ipynb-->first_author_race_pbm_p1_vs_p2.png
extract_funding.py-->covid_data.csv
extract_funding.py-->funding_data.tsv
data_visualizations_bl.ipynb-->animal_score.png
data_visualizations_bl.ipynb-->human_score.png
data_visualizations_bl.ipynb-->molecular_cellular_score.png
This is a list of search terms that we will use for testing. Comes from a google doc not linked here because I'm not sure we want it public.
Fields:
Query- query string used to search for publicationsCategory- Rare diseases | Signaling pathways | Autoimmune diseases | Cells | Infectious bacteria | Medical devices | Drugs | Social determinants of health, health equitiesSource- link to source of term
Below is a visualization of the query term spread:
This is a list of all the pmids for the terms we're interested in. Here's what the fields mean:
pmid- PubMed IDquery- query string, read in from ./data/in/team1_search_strats_search_term+.csv (from Travis' google doc)search_type- relevant | pubdate_descpage- 1 | 2
Columns of data pulled directly from the PubMed API or iCite:
iCite columns
relative_citation_ratio, RCR measure of influencehuman, Prediction from MeSH on human subjectanimal, Prediction from MeSH on animal subjectsmolecular_cellular, Prediction from MeSH on cellular subjectsapt, Approx. potential to clinically translateis_clinical, Bool, clinical or guidelinecitation_count, Raw of citationscited_by, PMIDs of citationsreferences, PMIDs of references
PubMed columns
titleabstractjournal- coded as 1 = US, England, or Ireland (UK), 0 = any other countryauthorsaffiliationspubdatemesh_termspublication_typeschemical_listkeywordslanguagescountry
Grant information is extracted from the pmid_xmls via extract_funding.py
pmidgrant_idgrant_acronymgrant_countrygrant_agency
A raw xml response from the pubmed api for each publication. Saved in the format of PMID.xml (e.g., 61455.xml). These aren't used in the analysis as pmid_data.csv has all the metadata in csv format. These may be used for confirmation.
This file isn't populated properly for books as it just loads article metadata. This is a small number of items (122 out of 7206 publications) so shouldn't impact results too much.
- readability_fk_score_abstract.csv - median flesch-kincaid score of publication abstract
- readability_fk_score_title_abstract_combined.csv - median flesh-kincaid score of publication title and abstract combined
- readability_fk_score_title.csv - median flesch-kincaid score of publication title
- author_count.csv - median number of authors per publication
- affiliation_count.csv - median number of affiliations per publication
- author_country.csv - most common country in page of search results for first authors
- author_affiliation.csv - most common affilitions in page of search results for first authors
Additional data processing in the Wiki
Here are the most statistically significant attributes from our analysis. For a full list of attribute visualizations, see visualization.md.
The full results for our search terms are provided here. Summarized results can be found in the table below:
| Feature | Comparison | p-value | Power | Direction |
|---|---|---|---|---|
| AIAN First Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 0.87 | Date |
| AIAN Last Author | Date Order Page 1 vs. Relevance Page 1 | 0.14 | 0.32 | Date |
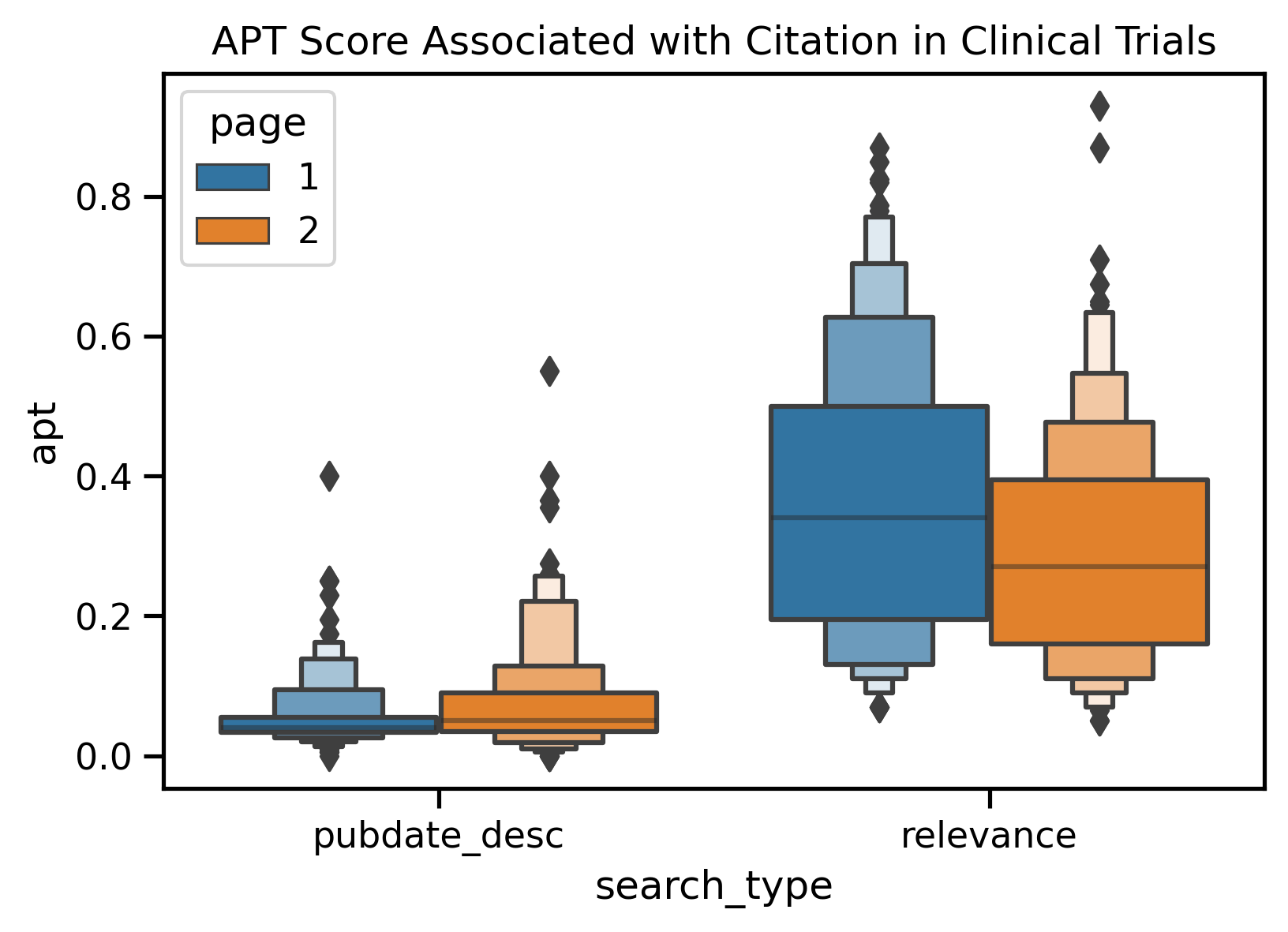
| APT Score | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
| APT Score | Relevance Page 1 vs. Relevance Page 2 | <0.01 | 0.93 | Page 1 |
| Affiliation Count | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Best Match |
| Affiliation Count | Relevance Page 1 vs. Relevance Page 2 | 0.23 | 0.22 | Page 1 |
| Animal Biomed Triangle | Date Order Page 1 vs. Relevance Page 1 | 0.19 | 0.26 | Best Match |
| Animal Biomed Triangle | Relevance Page 1 vs. Relevance Page 2 | 0.78 | 0.06 | Page 2 |
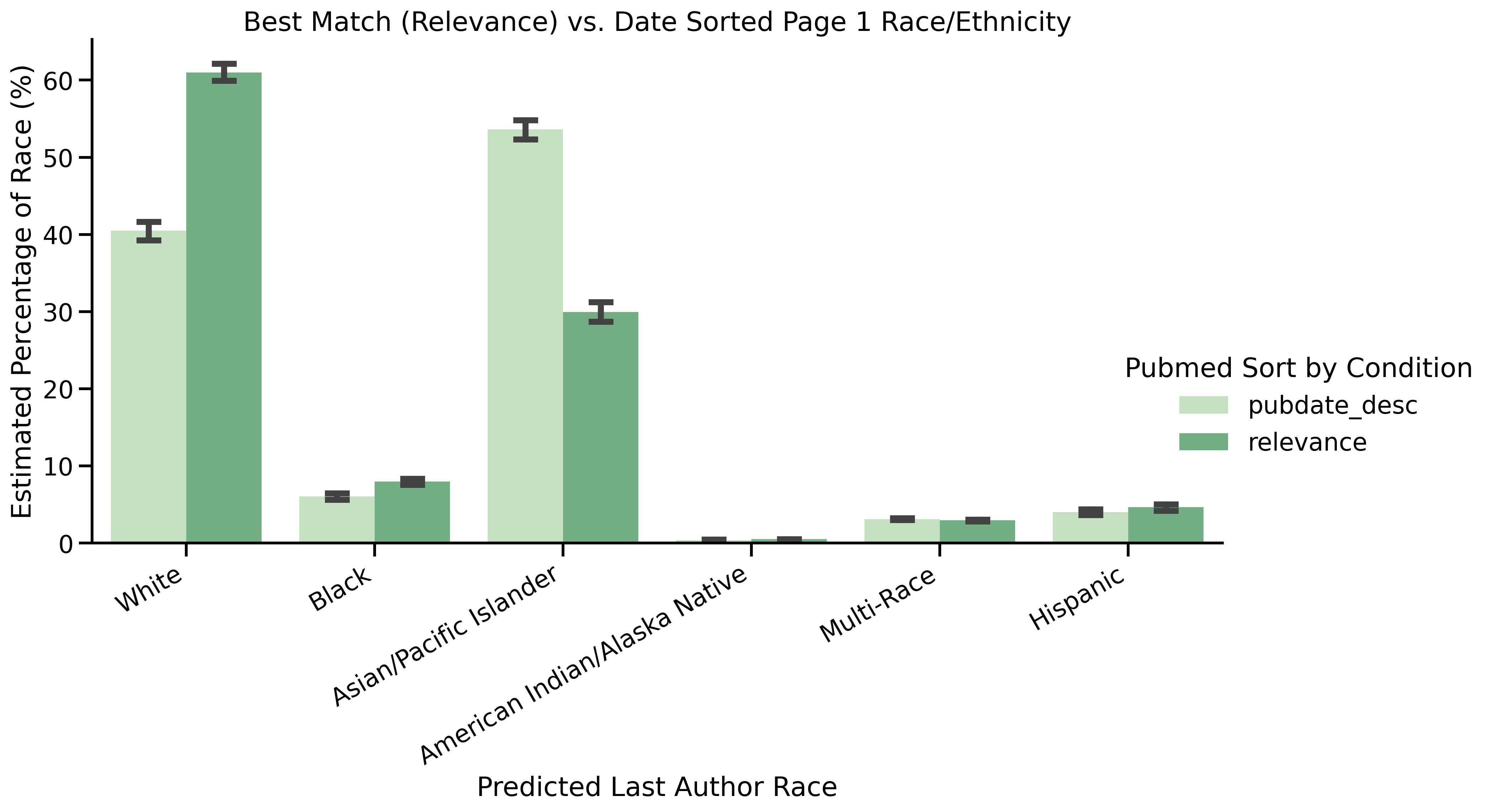
| Asian First Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Best Match |
| Asian Last Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Best Match |
| Author Count | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Best Match |
| Author Count | Relevance Page 1 vs. Relevance Page 2 | 0.49 | 0.11 | Page 2 |
| Black First Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
| Black Last Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 0.91 | Date |
| Country of Origin of Journal | Date Order Page 1 vs. Relevance Page 1 | 0.91 | 0.05 | Best Match |
| Country of Origin of Journal | Relevance Page 1 vs. Relevance Page 2 | 0.97 | 0.05 | Page 2 |
| FK Readability Abstract | Date Order Page 1 vs. Relevance Page 1 | 0.03 | 0.56 | Best Match |
| FK Readability Abstract | Relevance Page 1 vs. Relevance Page 2 | 0.45 | 0.12 | Page 2 |
| FK Readability Title | Date Order Page 1 vs. Relevance Page 1 | 0.17 | 0.28 | Best Match |
| FK Readability Title | Relevance Page 1 vs. Relevance Page 2 | 0.38 | 0.14 | Page 2 |
| First Author Gender | Date Order Page 1 vs. Relevance Page 1 | 0.03 | 0.60 | Best Match |
| First Author Gender | Relevance Page 1 vs. Relevance Page 2 | 0.01 | 0.76 | Page 1 |
| Hispanic First Author | Date Order Page 1 vs. Relevance Page 1 | 0.92 | 0.05 | Date |
| Hispanic Last Author | Date Order Page 1 vs. Relevance Page 1 | 0.26 | 0.20 | Date |
| Human Biomed Triangle | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 0.83 | Date |
| Human Biomed Triangle | Relevance Page 1 vs. Relevance Page 2 | 0.71 | 0.07 | Page 1 |
| Is English Only | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Best Match |
| Is English Only | Relevance Page 1 vs. Relevance Page 2 | 0.18 | 0.26 | Page 1 |
| Last Author Gender | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 0.93 | Best Match |
| Last Author Gender | Relevance Page 1 vs. Relevance Page 2 | 0.10 | 0.37 | Page 2 |
| Mol_Cell Biomed Triangle | Date Order Page 1 vs. Relevance Page 1 | 0.02 | 0.63 | Best Match |
| Mol_Cell Biomed Triangle | Relevance Page 1 vs. Relevance Page 2 | 0.86 | 0.05 | Page 2 |
| Multi-Race First Author | Date Order Page 1 vs. Relevance Page 1 | 0.17 | 0.27 | Best Match |
| Multi-Race Last Author | Date Order Page 1 vs. Relevance Page 1 | 0.24 | 0.22 | Best Match |
| Other Funding | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
| Other Funding | Relevance Page 1 vs. Relevance Page 2 | 0.45 | 0.12 | Page 1 |
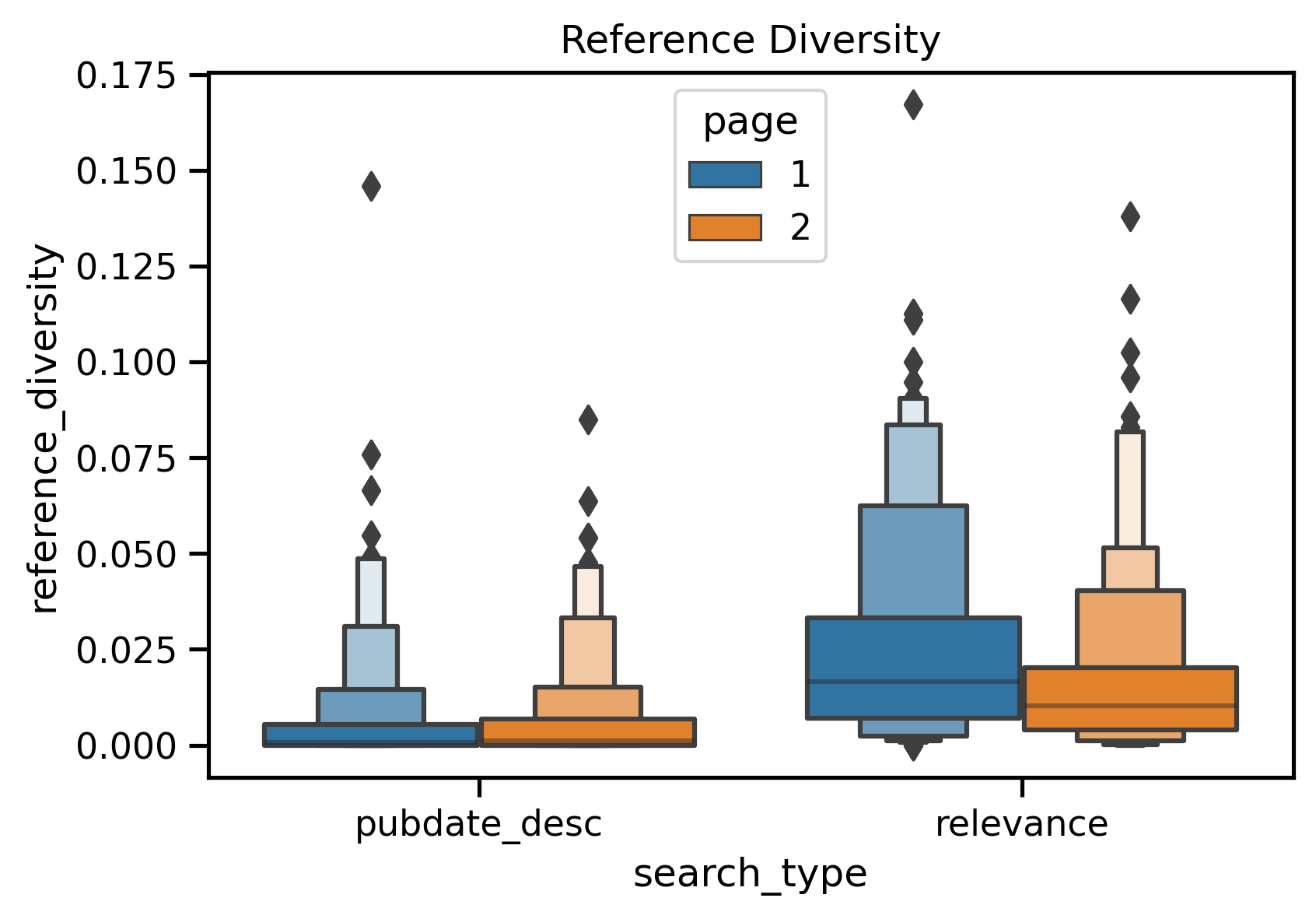
| Reference Diversity | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
| Reference Diversity | Relevance Page 1 vs. Relevance Page 2 | <0.01 | 0.84 | Page 1 |
| Relative Citation Ratio | Date Order Page 1 vs. Relevance Page 1 | 0.10 | 0.16 | Date |
| Relative Citation Ratio | Relevance Page 1 vs. Relevance Page 2 | <0.01 | 0.99 | Page 1 |
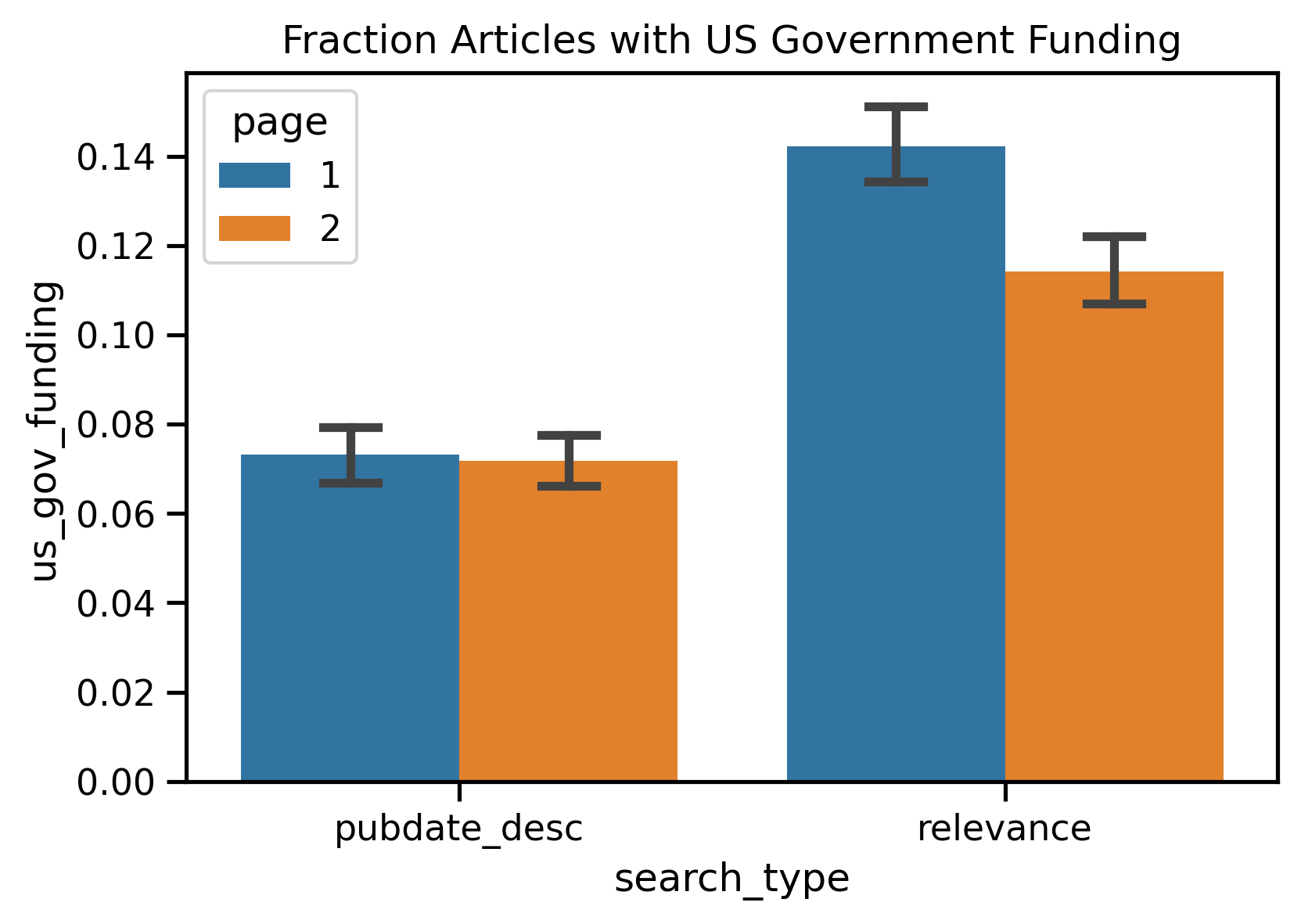
| US Gov Funding | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
| US Gov Funding | Relevance Page 1 vs. Relevance Page 2 | 0.01 | 0.71 | Page 1 |
| White First Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
| White Last Author | Date Order Page 1 vs. Relevance Page 1 | <0.01 | 1.00 | Date |
It is important to detect and mitigate bias in search and retrieval algorithms in order to achieve the three pillars for fairness - transparency, impartiality, and inclusion [5]. Systems continue to be biased as long as the data they receive is biased. Fairness via AI is based on the assertion that AI can be used to help “detect, mitigate, and remedy situations that are inherently unequal, unjust and unfair in society"[2]. Obermeyer et al [6] offer these strategies to mitigate bias:
- Document algorithms using transparent methods such as including goal, training process and performance
- Set up protocols to mitigate bias by providing a pathway for users to report bias, identification bias
- Establish an ongoing team to oversee bias mitigation measures
Continously training and re-training the learning models using representative and more accurate user search and click through data (as well as collabrative filtering) can help continous detection and elimination of biases. The measures of bias may also need to be defined more clearly and operationalized in future research and implementation.
These insights and their significance to useful for PubMed stakeholders including: users, PubMed developers, Information Retrieval researchers.
The Best Match algorithm is known to prefer reviews. We hypothesize that this may be driving some of the larger correlations like race and government funding. These insights are derived from our best judgement and the limited time we had to consider the results during the Codeathon.
It seems like RCR score and reference diversity is being driven by a "rich-get-richer" network effort, while APT may be driven by the types of users the data was trained off of (with a preference towards to clinical users).
The limitations of this work to be finished in a later study include:
- Expanding the breadth of search terms
- Fixing parsing issues (such as books vs articles)
- Systematic study of correlation of review articles to determine causality
- Using a control of most recent sort, this suffers from incomplete information from PubMed
- Fiorini N, Canese K, Starchenko G, Kireev E, Kim W, Miller V, Osipov M, Kholodov M, Ismagilov R, Mohan S, Ostell J, Lu Z. Best Match: New relevance search for PubMed. PLoS Biol. 2018 Aug 28;16(8):e2005343. doi: 10.1371/journal.pbio.2005343. PMID: 30153250; PMCID: PMC6112631. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6112631/
- Dori-Hacohen, S., Montenegro, R., Murai, F., Hale, S., Sung, K., Blain, M., & Edwards-Johnson, J. (2021). Fairness via AI: Bias Reduction in Medical Information. ArXiv, abs/2109.02202. Available at: https://www.researchgate.net/publication/354400541_Fairness_via_AI_Bias_Reduction_in_Medical_Information
- Morik, M., Singh, A., Hong, J., & Joachims, T. (2020). Controlling Fairness and Bias in Dynamic Learning-to-Rank. Proceedings of the 43rd International ACM SIGIR Conference on Research and Development in Information Retrieval. Available at: https://arxiv.org/pdf/2005.14713.pdf
- Kiester, Lucy & Turp, Clara. (2022). Artificial intelligence behind the scenes: PubMed’s Best Match algorithm. Journal of the Medical Library Association. 110. 10.5195/jmla.2022.1236. Available at: https://jmla.pitt.edu/ojs/jmla/article/view/1236
- Lever, J., Gakkhar, S., Gottlieb, M., Rashnavadi, T., Lin, S., Siu, C., Smith, M., Jones, M. R., Krzywinski, M., Jones, S., & Wren, J. (2018). A collaborative filtering-based approach to biomedical knowledge discovery. Bioinformatics (Oxford, England), 34(4), 652–659. https://doi.org/10.1093/bioinformatics/btx613
- Obermeyer, Z., Nissan, R., Stern, M., Eaneff, S., Bembeneck, E. J., & Mullainathan, S. (2021). Algorithmic Bias Playbook. Center for Applied AI at Chicago Booth. Available at: https://www.ftc.gov/system/files/documents/public_events/1582978/algorithmic-bias-playbook.pdf
Thanks to Team4 and @Danizen for their pubmed api code that we used to pull down articles.
- Shruthi Chari (data lead)
- Travis Hoppe (project lead)
- Alex Sticco
- Alexa M. Salsbury
- Ben Rogers
- Brian Lee
- Brooks Leitner
- Caroline Trier
- Linda M. Hartman
- Preeti G. Kochar
- Sridhar Papagari Sangareddy