Simple API for Python Integration with NCBI .
Explore the docs »
View Demo
·
Report Bug
·
Request Feature
Here's Why PyNCBI 🧬 :
When working with methylation data, NCBI might be one of the most extensive open source databases that provide the methylation data and the information around it. When working with NCBI on a day-to-day basis, searching, querying, and extracting information may prove to be a time-consuming and headache-producing challenge. PyNCBI strives to answer all needs a researcher might need when communicating with NCBI using a straightforward python API that allows to quickly test, extract, analyze and download relevant data.
pip install PyNCBI
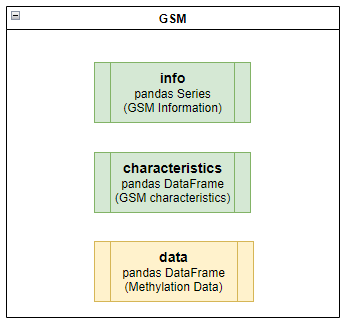
The GSM API extracts all info from a GSM card and downloads the methylation data, and renders the beta values ready for work. After extracting and preprocessing the data once, that GSM instance will be cached for your convenience; each following time, you will reference the same GSM id the cached version will be loaded. The GSM class contains the following attributes:
- array_type - the array type used to sequence the data
- gse - the parent GSM id
- info - a Pandas Series contacting the entire GSM card information
- data - a Pandas DataFrame containing the probes and matching beta values
- characteristics - the parsed characteristics section from the GSM info section
from PyNCBI import GSM
# Build and populate with data an instance of a GSM container
example_gsm = GSM('GSM1518180')
print(example_gsm)Output:
GSM: GSM1518180 | GSE: GSE62003
tissue: Whole blood
Sex: Male
age: 77
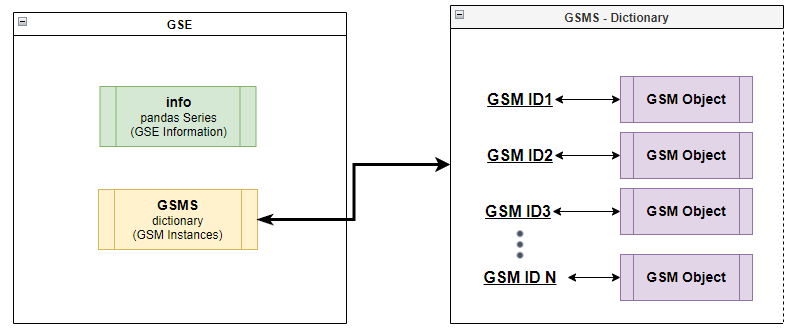
Create a GSE class instance that contains all the information on a given GSE id, the instance is populated with instances of the GSM class that cointains the methylation data and information for each GSM in the given GSE. The GSE class contains the following attributes:
- info - a Pandas Series contacting the entire GSE card information
- GSMS - a dictionary, string ('gsm id') to a GSM object instance reference
from PyNCBI import GSE
# Build and populate with data an instance of a GSE container
example_gse = GSE('GSE85506',mode='supp')
print(example_gse)Output:
GSE: GSE85506
Array Type: GPL13534 (450k)
Number of Samples: 47
Title: DNA methylation analysis in women with fibromyalgia
Inside each GSE object is a dictionary of GSM objects Example:
example_gse["GSM2267972"]
# the above returns the GSM obeject that matches the id given i.e "GSM2267972"Output:
GSM: GSM2267972 | GSE: GSE85506
tissue: peripheral blood
gender: female
group: case
age: 56
inhibition (average values): 1.586085
facilitation values (average): 2.60410125
- GSE Wise Card Information Extraction
- GSM Card Information Extraction
- GSE Wise Methylation Data Extraction
- GSM Card Methylation Data Extraction
- IDAT File Parsing Management Based on methylprep
- Single GSM API
- Single GSE API
See the open issues for a list of proposed features (and known issues).
Contributions are what make the open-source community such a powerful place to create new ideas, inspire, and make progress. Any contributions you make are greatly appreciated.
- Fork the Project
- Create your Feature Branch (
git checkout -b feature/AmazingFeature) - Commit your changes (
git commit -m 'Add some AmazingFeature') - Push to the Branch (
git push origin feature/AmazingFeature) - Open a Pull Request
Distributed under the MIT license. See LICENSE for more information.
Thomas Konstantinovsky - [email protected]
Project Link: https://github.com/MuteJester/PyNCBI