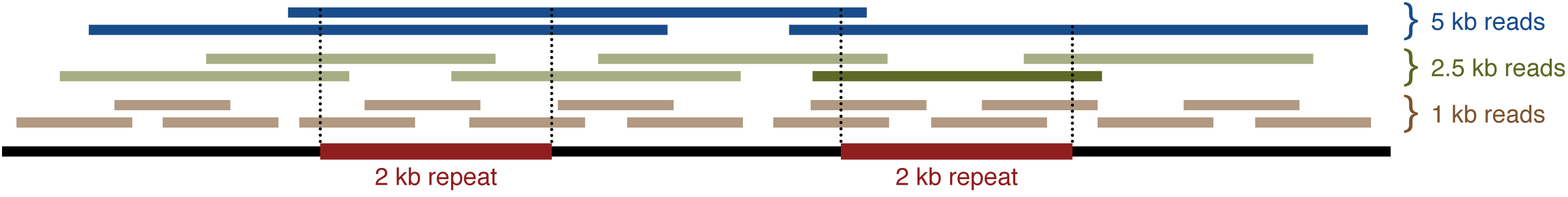Unicycler is a hybrid assembly pipeline for bacterial genomes. It uses both Illumina reads and long reads (PacBio or Nanopore) to produce complete and accurate assemblies.
- Introduction
- Requirements
- Installation
- Quick usage
- How it works
- Conservative, normal and bold
- Options and usage
- Output files
- Tips
- Unicycler align
- Unicycler polish
- Citation
- Acknowledgements
- License
As input, Unicycler takes a good set of Illumina reads from a bacterial isolate (required) and long reads from the same isolate (optional). If the input is sufficient, it will produce a completed assembly of circularised sequences.
Reasons to use Unicycler:
- It has very low misassembly rates.
- It can cope with very repetitive genomes, such as Shigella.
- It correctly handles plasmids of varying depth.
- It works with long reads of any quality – even Nanopore reads classed as 'fail' can be used as input.
- It works with any long read depth. 10x or more may be required to complete a genome, but Unicycler can make nearly-complete genomes with far fewer long reads.
- Even if you have no long reads, it functions as a SPAdes optimiser and produces very good Illumina assemblies.
- It produces an assembly graph in addition to a contigs FASTA file, viewable in Bandage.
- It's easy to use, runs with just one command and doesn't require tinkering with parameters!
Reasons to not use Unicycler:
- You only have long reads, not Illumina reads (try Canu instead).
- Your Illumina reads are poor quality (Unicycler requires a good short read assembly graph – more info here).
- You're assembling a large eukaryotic genome or a metagenome (Unicycler is designed for small genomes like bacterial isolates).
- Your Illumina reads and long reads are from different isolates.
- You're very impatient (Unicycler is not as fast as alternatives).
- Linux or macOS
- Python 3.4 or later
- C++ compiler
- SPAdes 3.6.2 or later
- setuptools (only required for installation of Unicycler)
Unicycler needs the following tools for certain parts of its pipeline. They are optional, but without them Unicycler will not be able to perform all tasks:
- Pilon (required for polishing)
- Java (required for polishing)
- Bowtie2 (required for polishing)
- Samtools version 1.0 or later (required for polishing)
- BLAST+ (required for rotating finished assemblies)
Bandage isn't required to run Unicycler, but it is very helpful for manually investigating assemblies (the graph images in this README were made with Bandage). Make sure to get the latest release for full compatibility with Unicycler.
These instructions install the most up-to-date version of Unicycler:
git clone https://github.com/rrwick/Unicycler.git
cd Unicycler
python3 setup.py installNotes:
- If the last command complains about permissions, you may need to run it with
sudo. - If you want a particular version of Unicycler, download the source from the releases page instead of cloning from GitHub.
- Install just for your user:
python3 setup.py install --user- If you get a strange 'can't combine user with prefix' error, read this.
- Install to a specific location:
python3 setup.py install --prefix=$HOME/.local - Install with pip (local copy):
pip3 install path/to/Unicycler - Install with pip (from GitHub):
pip3 install git+https://github.com/rrwick/Unicycler.git - Install with specific Makefile options:
python3 setup.py install --makeargs "CXX=icpc"
This approach compiles Unicycler code, but doesn't copy executables anywhere:
git clone https://github.com/rrwick/Unicycler.git
cd Unicycler
makeNow instead of running unicycler, you instead use path/to/unicycler-runner.py.
These commands use the reads you'll find in the sample_data directory. They are synthetic reads generated from plasmids in a Shigella sonnei reference.
Short read-only assembly:
unicycler -1 short_reads_1.fastq.gz -2 short_reads_2.fastq.gz -o output_dir
Hybrid assembly:
unicycler -1 short_reads_1.fastq.gz -2 short_reads_2.fastq.gz -l long_reads_high_depth.fastq.gz -o output_dir
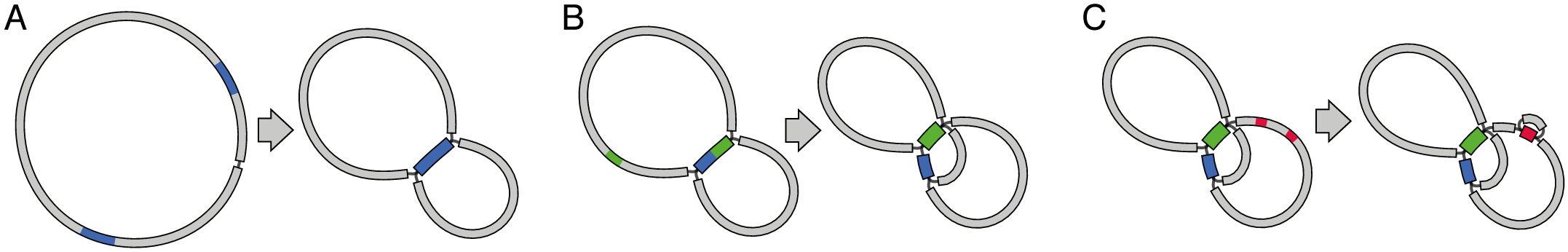
To understand what Unicycler is doing, you need to know about assembly graphs. For a thorough introduction, I'd suggest this tutorial or the Velvet paper. But in short, an assembly graph is a data structure where contigs aren't disconnected sequences but can have connections to each other:
Just contigs: Assembly graph:
TCGAAACTTGACGCGAGTCGC CTTGTTTA
TGCTACTGCTTGATGATGCGG / \
TGTCCATT TCGAAACTTGACGCGAGTCGC TGCTACTGCTTGATGATGCGG
CTTGTTTA \ /
TGTCCATT
Most assemblers use graphs internally to produce their assemblies, but users often ignore the graph in favour of the conceptually simpler FASTA file of contigs. When a genome assembly is 100% complete, we have one contig per chromosome/plasmid and there's no real need for the graph. But most assemblies are not complete (especially short read assemblies), and a graph can describe an incomplete assembly much better than contigs alone.
The main reason we can't get a complete assembly from short reads is that DNA usually contains repeats – the same sequence occurring two or more times in the genome. When a repeat is longer than the reads (or for paired-end sequencing, longer than the insert size), it forms a single contig in the assembly graph with multiple connections in and multiple connections out.
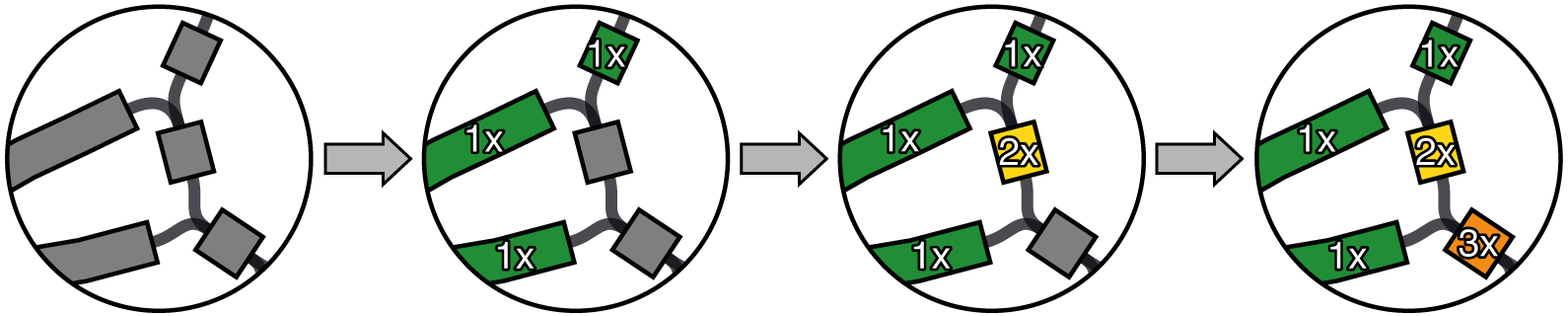
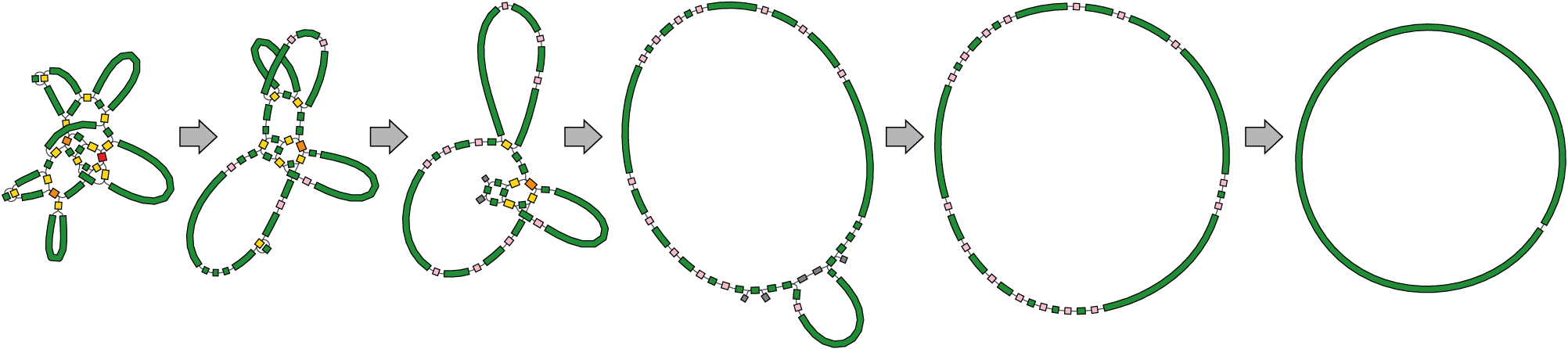
Here is what happens to a simple bacterial assembly graph as you add repeats to the genome:
As repeats are added, the graph becomes increasingly tangled (and real assembly graphs get a lot more tangled than that).
To complete a bacterial genome assembly (i.e. find the one correct sequence for each chromosome/plasmid), we need to resolve the repeats. This means finding which way into a repeat matches up with which way out. Short reads don't have enough information for this but long reads do.
Assembly graphs come in many different varieties, but we are particularly interested in the kind produced by SPAdes, because that is what Unicycler uses.
SPAdes graphs are made by performing a De Bruijn graph assembly with a range of different k-mer sizes, from small to large (see the SPAdes paper). Each assembly builds on the previous one, which allows SPAdes to get the advantages of both small k-mer assemblies (a more connected graph) and large k-mer assemblies (ability to resolve repeats). As a consequence of how SPAdes combines k-mers, two contigs in a SPAdes graph that connect will overlap by their k-mer size (more info on the Bandage wiki page).
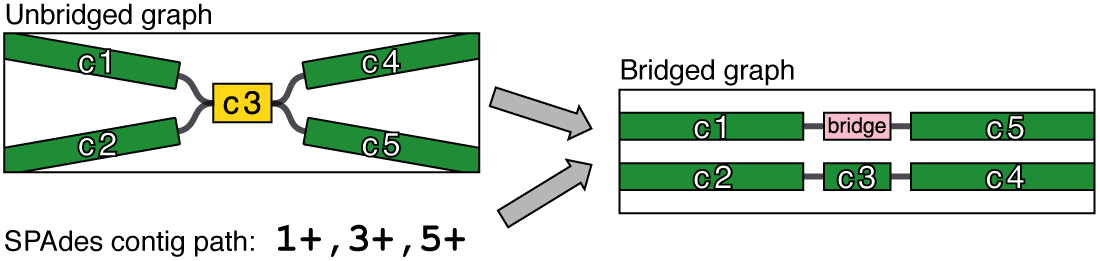
After producing the graph, SPAdes can perform further repeat resolution by using paired-end information. Since two reads in a pair are close to each other in the original DNA, SPAdes can use this to trace paths in the graph to form larger contigs (see their paper on ExSPAnder). However, the SPAdes contigs with repeat resolution do not come in graph form – they are only available in a FASTA file.
Unicycler uses SPAdes to get an assembly graph made from Illumina reads. Since Illumina reads are accurate, this graph has very few mistakes. But since Illumina reads are short, it will also contain unresolved repeats. Unicycler then uses the SPAdes repeat resolution and long read alignments to build 'bridges' between non-repeat contigs, simplifying the graph into fewer, longer contigs.
Essentially, Unicycler is a scaffolder which uses long reads to properly arrange Illumina contigs. But unlike a naive scaffolding tool which operates on assembled contigs, Unicycler works on an assembly graph. This gives it much more information to complete assemblies and a lower risk of mistakes.
Here is the process in a bit more detail:
Unicycler uses SPAdes' built-in read correction step before assembling the Illumina reads. This can be disabled with --no_correct if your Illumina reads are very high quality or you've already performed read QC.
Unicycler uses SPAdes to assemble the Illumina reads into an assembly graph. It tries assemblies at a wide range of k-mer sizes, evaluating the graph at each one. It chooses the graph which best minimises both contig count and dead end count. If the Illumina reads are good, it produces an assembly graph with long contigs but few to no dead ends (more info here). Since a typical bacterial genome has no dead ends (the sequences are circular) an ideal assembly graph won't either.
A raw SPAdes graph can also contain some 'junk' sequences due to sequencer artefacts or contamination, so Unicycler performs some graph cleaning to remove these. Therefore, small amounts of contamination in the Illumina reads should not be a problem.
In order to scaffold the graph, Unicycler must distinguish between single copy contigs and repeats. It does this with a greedy algorithm that uses both read depth and graph connectivity:
This process does not assume that all single copy contigs have the same read depth, which allows it to identify single copy contigs from plasmids as well as the chromosome.
At this point, the assembly graph does not contain the SPAdes repeat resolution. To apply this to the graph, Unicycler builds bridges between single copy contigs using the information in the SPAdes contigs.paths file. These are applied to the graph to make the short_read_bridges_applied.gfa output – the most resolved graph Unicycler can make using only the Illumina reads.
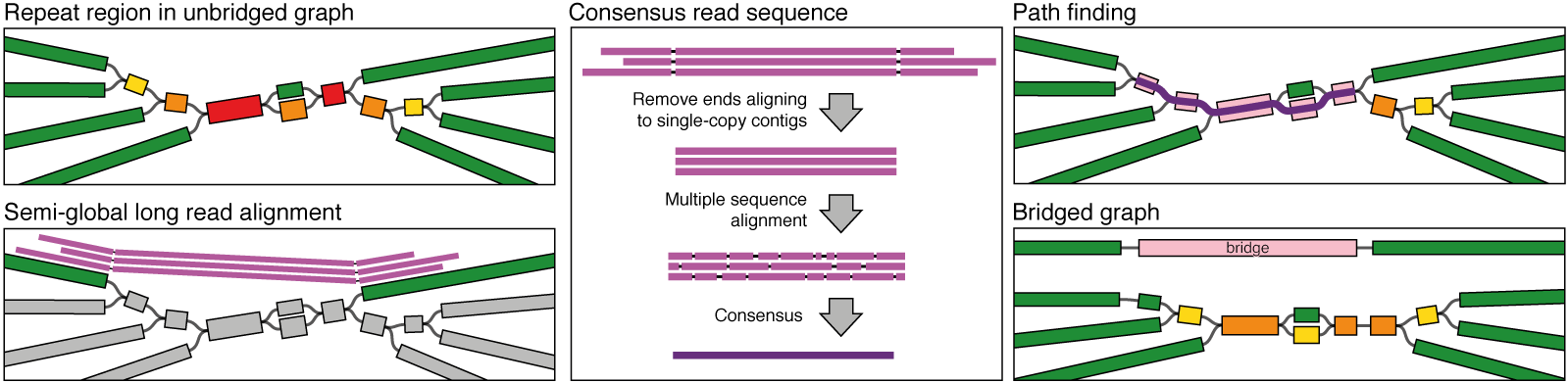
Long reads are the most useful source of information for resolving the assembly graph, so Unicycler semi-globally aligns them to the contigs (see Unicycler align for more information). For each pair of single copy contigs which are linked by read alignments, Unicycler uses the read consensus sequence to find a connecting path and creates a bridge.
At this point of the pipeline there can be many bridges, some of which may conflict. Unicycler therefore assigns a quality score to each based on all available evidence (e.g. read alignment quality, graph path match, read depth consistency). Bridges are then applied in order of decreasing quality so whenever there is a conflict, only the most supported bridge is used. A minimum quality threshold prevents the application of low evidence bridges (see Conservative, normal and bold for more information).
If the above steps have resulted in any simple, circular sequences, then Unicycler will attempt to rotate/flip them to begin at a consistent starting gene. By default this is dnaA or repA, but users can specify their own with the --start_genes option.
Finally, Unicycler does a single pass of short read polishing using Pilon. For a more extensive polishing process, see the section on Unicycler polish
Unicycler can be run in three modes: conservative, normal (the default) and bold, set with the --mode option. Conservative mode is least likely to produce a complete assembly but has a very low risk of misassembly. Bold mode is most likely to produce a complete assembly but carries greater risk of misassembly. Normal mode is intermediate regarding both completeness and misassembly risk.
If the structural accuracy of your assembly is paramount to your research, conservative mode is recommended. If you want a completed genome, even if it contains a mistake or two, then use bold mode.
The specific differences between the three modes are as follows:
| Mode | Invocation | Short read bridges | Bridge quality threshold | Contig merging |
|---|---|---|---|---|
| conservative | ‑‑mode conservative |
not used | high (25) | contigs are only merged with bridges |
| normal | ‑‑mode normal(or nothing) |
used | medium (10) | contigs are merged with bridges and when their multiplicity is 1 |
| bold | ‑‑mode bold |
used | low (1) | contigs are merged wherever possible |
In the above example, the conservative assembly is incomplete because some bridges fell below the quality threshold and were not applied. Its contigs, however, are extremely reliable. Normal mode nearly gave a complete assembly, but a couple of unmerged contigs remain. Bold mode completed the assembly, but since lower confidence regions were bridged and merged, there is a larger risk of error.
Run unicycler --help to view the program's most commonly used options:
usage: unicycler-runner.py [-h] [--help_all] [--version] -1 SHORT1 -2 SHORT2 [-s UNPAIRED] [-l LONG] -o OUT
[--verbosity VERBOSITY] [--min_fasta_length MIN_FASTA_LENGTH] [--keep KEEP] [-t THREADS]
[--mode {conservative,normal,bold}] [--linear_seqs LINEAR_SEQS]
__
\ \___
\ ___\
//
____// _ _ _ _
//_ //\\ | | | | |_| | |
// \// \\ | | | | _ __ _ ___ _ _ ___ | | ___ _ __
|| (O) || | | | || '_ \ | | / __|| | | | / __|| | / _ \| '__|
\\ \_ // | |__| || | | || || (__ | |_| || (__ | || __/| |
\\_____// \____/ |_| |_||_| \___| \__, | \___||_| \___||_|
__/ |
|___/
Unicycler: a hybrid assembly pipeline for bacterial genomes
Help:
-h, --help Show this help message and exit
--help_all Show a help message with all program options
--version Show Unicycler's version number
Input:
-1 SHORT1, --short1 SHORT1 FASTQ file of first short reads in each pair (required)
-2 SHORT2, --short2 SHORT2 FASTQ file of second short reads in each pair (required)
-s UNPAIRED, --unpaired UNPAIRED FASTQ file of unpaired short reads (optional)
-l LONG, --long LONG FASTQ or FASTA file of long reads (optional)
Output:
-o OUT, --out OUT Output directory (required)
--verbosity VERBOSITY Level of stdout and log file information (default: 1)
0 = no stdout, 1 = basic progress indicators, 2 = extra info,
3 = debugging info
--min_fasta_length MIN_FASTA_LENGTH Exclude contigs from the FASTA file which are shorter than this length
(default: 1)
--keep KEEP Level of file retention (default: 1)
0 = only keep final files: assembly (FASTA, GFA and log),
1 = also save graphs at main checkpoints,
2 = also keep SAM (enables fast rerun in different mode),
3 = keep all temp files and save all graphs (for debugging)
Other:
-t THREADS, --threads THREADS Number of threads used (default: 8)
--mode {conservative,normal,bold} Bridging mode (default: normal)
conservative = smaller contigs, lowest misassembly rate
normal = moderate contig size and misassembly rate
bold = longest contigs, higher misassembly rate
--linear_seqs LINEAR_SEQS The expected number of linear (i.e. non-circular) sequences in the
underlying sequence (default: 0)
Run unicycler --help_all to see a complete list of the program's options. These allow you to turn off parts of the pipeline, specify the location of tools (only necessary if they are not in PATH) and adjust various settings:
SPAdes assembly:
These options control the short read SPAdes assembly at the beginning of the Unicycler pipeline.
--spades_path SPADES_PATH Path to the SPAdes executable (default: spades.py)
--no_correct Skip SPAdes error correction step (default: conduct SPAdes error correction)
--min_kmer_frac MIN_KMER_FRAC Lowest k-mer size for SPAdes assembly, expressed as a fraction of the read
length (default: 0.2)
--max_kmer_frac MAX_KMER_FRAC Highest k-mer size for SPAdes assembly, expressed as a fraction of the read
length (default: 0.95)
--kmer_count KMER_COUNT Number of k-mer steps to use in SPAdes assembly (default: 10)
Assembly rotation:
These options control the rotation of completed circular sequence near the end of the Unicycler pipeline.
--no_rotate Do not rotate completed replicons to start at a standard gene (default:
completed replicons are rotated)
--start_genes START_GENES FASTA file of genes for start point of rotated replicons (default:
start_genes.fasta)
--start_gene_id START_GENE_ID The minimum required BLAST percent identity for a start gene search
(default: 90.0)
--start_gene_cov START_GENE_COV The minimum required BLAST percent coverage for a start gene search
(default: 95.0)
--makeblastdb_path MAKEBLASTDB_PATH Path to the makeblastdb executable (default: makeblastdb)
--tblastn_path TBLASTN_PATH Path to the tblastn executable (default: tblastn)
Pilon polishing:
These options control the final assembly polish using Pilon at the end of the Unicycler pipeline.
--no_pilon Do not use Pilon to polish the final assembly (default: Pilon is used)
--bowtie2_path BOWTIE2_PATH Path to the bowtie2 executable (default: bowtie2)
--bowtie2_build_path BOWTIE2_BUILD_PATH
Path to the bowtie2_build executable (default: bowtie2-build)
--samtools_path SAMTOOLS_PATH Path to the samtools executable (default: samtools)
--pilon_path PILON_PATH Path to a Pilon executable or the Pilon Java archive file (default: pilon)
--java_path JAVA_PATH Path to the java executable (default: java)
--min_polish_size MIN_POLISH_SIZE Contigs shorter than this value (bp) will not be polished using Pilon
(default: 10000)
Graph cleaning:
These options control the removal of small leftover sequences after bridging is complete.
--min_component_size MIN_COMPONENT_SIZE
Unbridged graph components smaller than this size (bp) will be removed from
the final graph (default: 1000)
--min_dead_end_size MIN_DEAD_END_SIZE
Graph dead ends smaller than this size (bp) will be removed from the final
graph (default: 1000)
Long read alignment:
These options control the alignment of long reads to the assembly graph.
--contamination CONTAMINATION FASTA file of known contamination in long reads
--scores SCORES Comma-delimited string of alignment scores: match, mismatch, gap open, gap
extend (default: 3,-6,-5,-2)
--low_score LOW_SCORE Score threshold - alignments below this are considered poor (default: set
threshold automatically)
Unicycler's most important output files are assembly.gfa, assembly.fasta and unicycler.log. These are produced by every Unicycler run. Which other files are saved to its output directory depends on the value of --keep:
--keep 0retains only the important files. Use this setting to save drive space.--keep 1(the default) also saves some intermediate graphs.--keep 2also retains the SAM file of long read alignments to the graph. This ensures that if you rerun Unicycler with the same output directory (for example changing the mode to conservative or bold) it will run faster because it does not have to repeat the alignment step.--keep 3retains all files and saves many intermediate graphs. This is for debugging purposes and uses a lot of space. Most users should probably avoid this setting.
All files and directories are described in the table below. Intermediate output files (everything except for assembly.gfa, assembly.fasta and unicycler.log) will be prefixed with a number so they are in chronological order.
| File | Description | --keep level |
|---|---|---|
| spades_assembly/ | directory containing all SPAdes files and each k-mer graph | 3 |
| unbridged_graph.gfa | short read assembly graph before any bridges have been applied | 1 |
| short_read_bridges_applied.gfa | SPAdes contig bridges applied, before any cleaning or merging | 1 |
| cleaned.gfa | redundant contigs removed from the graph | 3 |
| merged.gfa | contigs merged together where possible | 3 |
| long_read_bridges_applied.gfa | long read bridges applied, before any cleaning or merging | 1 |
| read_alignment/ | directory containing long_read_alignments.sam |
2 |
| cleaned.gfa | redundant contigs removed from the graph | 3 |
| merged.gfa | contigs merged together where possible | 3 |
| final_clean.gfa | more redundant contigs removed | 1 |
| rotated.gfa | circular replicons rotated and/or flipped to a start position | 1 |
| polished.gfa | after a round of Pilon polishing | 1 |
| assembly.gfa | final assembly in GFA v1 graph format | 0 |
| assembly.fasta | final assembly in FASTA format (same contigs as in assembly.gfa) | 0 |
| unicycler.log | Unicycler log file (same info as stdout) | 0 |
Unicycler is thorough and accurate, but not particularly fast. In particular, the long read bridging step of the pipeline can take a while to complete. Two main factors influence the running time: the number of long reads (more reads take longer to align) and the genome size/complexity (finding bridge paths is more difficult in complex graphs).
Unicycler may only take an hour or so to assemble a small, simple genome with low depth long reads. On the other hand, a complex genome with many long reads may take 12 hours to finish or more. If you have a very high depth of long reads, you can make Unicycler run faster by subsampling for only the longest reads.
Using a lot of threads (with the --threads option) can make Unicycler run faster too. It will only use up to 8 threads by default, but if you're running it on a big machine with lots of CPU and RAM, feel free to use more!
The length of a long read is very important, typically more than its accuracy, because longer reads are more likely to align to multiple single copy contigs, allowing Unicycler to build bridges.
Consider a sequence with a 2 kb repeat:
In order to resolve the repeat, a read must span it by aligning to some sequence on either side. In this example, the 1 kb reads are shorter than the repeat and are useless. The 2.5 kb reads can resolve the repeat, but they have to be in just the right place to do so. Only one out of the six in this example is useful. The 5 kb reads, however, have a much easier time spanning the repeat and all three are useful.
So how long must your reads be for Unicycler to complete an assembly? Longer than the longest repeat in the genome. Depending on the genome, that might be a 1 kb insertion sequence, a 6 kb rRNA operon or a 50 kb prophage. If your reads are just a bit longer than the longest repeat, you'll probably need a lot of them. If they are much longer, then fewer reads should suffice. But in any scenario, longer is better!
Poretools can turn your Nanopore FAST5 reads into a FASTQ file appropriate for Unicycler. Here's an example command:
poretools fastq --type best --min-length 1000 path/to/fast5/dir/ > nanopore_reads.fastqIf you have 2D reads, --type best makes Poretools give only one FASTQ read per FAST5 file (if you have 1D reads, you can exclude that option). Adjust the --min-length 1000 parameter to suit your dataset – a larger value would be appropriate if you have lots of long reads.
Since Unicycler can tolerate low accuracy reads, Oxford Nanopore 1D sequencing is preferable to 2D, as it can provide twice as many reads. Unicycler will of course work with 2D reads, but you can get more out of your flow cell with 1D.
Unicycler needs decent Illumina reads as input – ideally with uniform read depth and 100% genome coverage.
You can look at the unbridged_graph.gfa file (the first graph Unicycler saves to file) in Bandage to get a quick impression of the Illumina read quality:
A is an very good Illumina read graph – the contigs are long and there are no dead ends. This read set is ideally suited for use in Unicycler.
B is also a good graph. The genome is more complex, resulting in a more tangled structure, but there are still very few dead ends (you can see one in the lower left). This read set would also work well in Unicycler.
C is a disaster! It is broken into many pieces, probably because parts of the genome got no read depth at all. While you can still use Unicycler to resolve this assembly with long reads, the risk of small errors and misassemblies is considerably higher. If you have sufficient long reads, I would instead recommend assembling the long reads with Canu and the polishing the resulting assembly using the Illumina reads and Pilon.
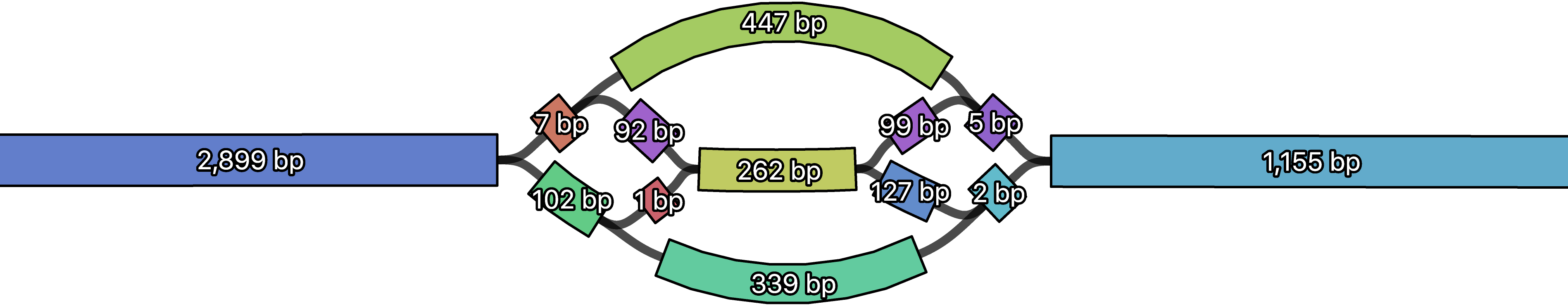
Confused by very small (e.g. 2 bp) contigs in Unicycler assemblies? Unlike a SPAdes graph where neighbouring sequences overlap by their k-mer size, Unicycler's final graph has no overlaps and the sequences adjoin directly. This means that contigs in complex regions can be quite short. They may be useless as stand-alone contigs but are still important in the graph structure.
If short contigs are a problem for your downstream analysis, you can use the --min_fasta_length to exclude them from Unicycler's FASTA file (they will still be included in the GFA file).
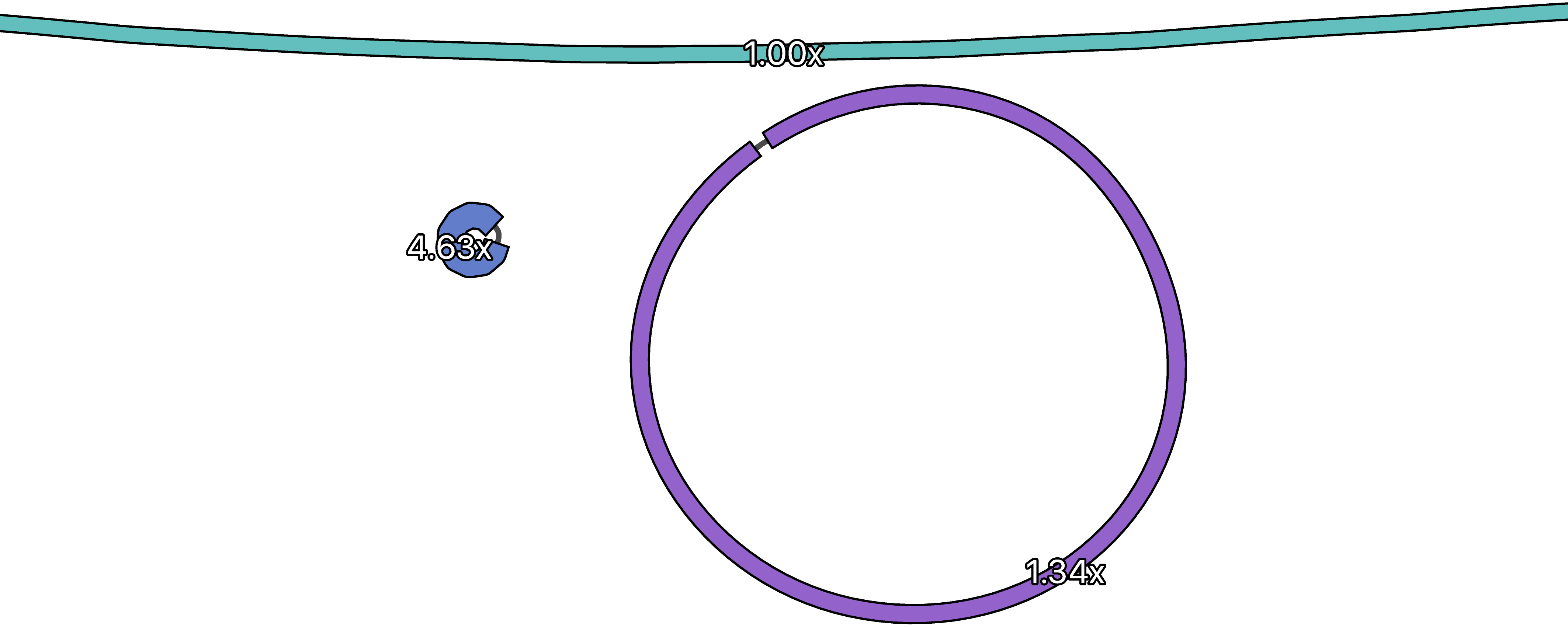
Unicycler normalises the depth of contigs in the graph to the median value. This typically means that the chromosome has a depth near 1x and plasmids may have different (typically higher) depths.
In the above graph, the chromosome is at the top (you can only see part of it) and there are two plasmids. The plasmid on the left occurs in approximately 4 or 5 copies per cell. For the larger plasmid on the right, most cells probably had one copy but some had more. Since sequencing biases can affect read depth, these per cell counts should be interpreted loosely.
If your long reads have known contamination, you can use the --contamination option to give Unicycler a FASTA file of the contaminant sequences. Unicycler will then discard any reads for which the best alignment is to the contaminant.
For example, if you've sequenced two isolates in succession on the same Nanopore flow cell, there may be residual reads from the first sample in the second run. In this case, you can supply a reference/assembly of the first sample to Unicycler when assembling the second sample.
Some Oxford Nanopore protocols include a lambda phage spike-in as a control. Since this is a common contaminant, you can simply use --contamination lambda to filter these out (no need to supply a FASTA file).
Unicycler's algorithm for sensitive semi-global alignment is available as a stand-alone alignment tool with the command unicycler_align.
Semi-global alignment (a.k.a. glocal, overlap or free end-gap alignment) will not clip an alignment until one of the two sequences ends. This can be where one sequence is contained within the other or where the two sequences overlap:
TAGAA GTGCCGGAACA GGCCACAC AGTAAGAT
||||| ||||||| ||||| |||||
ACTAGAACG GCCGGAA GGCTGGCCA AAGATCTTG
In contrast, local alignment will align only the best matching parts, clipping the alignment where the quality becomes poor:
CGAACAGCATACTTG
||||||||
ACGTCAGACTCAGCATACGCATCTAGA
Semi-global alignment is appropriate when there are no structural differences between the query and reference sequences. For example, when you have a short read assembly graph and long reads from the same bacterial isolate (as is the case in the Unicycler pipeline). In this scenario, there may be small scale differences (due to read errors) but no large scale differences, and semi-global alignment is ideal.
Semi-global alignment is probably not appropriate for mapping reads to a more distant reference genome. It does not cope with points of structural variation between the sample and the reference. For example, if the sample had a deletion relative to the reference, a read spanning that deletion would align poorly with semi-global alignment:
read: AACACTAAACTTAGTCCCAA
||||||||||| | | |
reference: GATCCCAACACTAAACTCTGGGGCGAACGGCGTAGTCCCAAGAGT
Local alignment (which can align only part of the read) would be more appropriate:
read: AACACTAAACT TAGTCCCAA
||||||||||| |||||||||
reference: GATCCCAACACTAAACTCTGGGGCGAACGGCGTAGTCCCAAGAGT
Try BWA-MEM, LAST or BLASR if you need a local alignment tool.
Regular alignment:
unicycler_align --reads queries.fastq --ref target.fasta --sam output.sam
Very sensitive (and slow) alignment:
unicycler_align --reads queries.fastq --ref target.fasta --sam output.sam --sensitivity_level 3
Setting some additional thresholds:
unicycler_align --reads queries.fastq --ref target.fasta --sam output.sam --min_len 1000 --low_score 80.0
Unicycler polish is a script to repeatedly polish a completed assembly using all available reads. It can be given Illumina reads, long reads or (ideally) both. When both Illumina and long reads are available, Unicycler polish can fix assembly errors, even in repetitive parts of the genome which cannot be polished by short reads alone.
- If polishing with Illumina reads: Pilon, Java, Bowtie2, Samtools (version 1.0 or later)
- If polishing with PacBio reads: pbalign, BLASR, GenomicConsensus
- PacBio software is most easily installed using pitchfork.
- If polishing with both Illumina and long reads (e.g. Nanopore): Racon, FreeBayes
Unicycler polish uses an exhaustive iterative process that is time-consuming but can be necessary to resolve the sequence in repeat regions. For example, consider a genome with two very similar regions, A and B, and there are assembly errors in both. Polishing is initially difficult because the errors may cause reads which should map to A to instead map to B and vice versa. However, after some of these errors are fixed, more reads will map to their correct locations, allowing for more errors to be fixes, allowing more reads to map correctly, etc.
- If Illumina reads are available:
- Run Pilon in 'bases' mode (substitutions and small indels). If any changes were suggested, apply them and repeat this step.
- Run Pilon in 'local' mode (larger variants), and assess each change with ALE. If any variant improves the ALE score, apply it and go back to step 1-i.
- If long reads are available:
- Run GenomicConsensus/Racon and gather all suggested small changes.
- Use FreeBayes to assess each long read-suggested change by looking for ambiguity in the Illumina read mapping. If any were found, apply them and go back to step 2-i.
- If Illumina reads are available:
- Execute step 1 again.
- Run Pilon/GenomicConsensus/Racon again (all that apply) and assess each suggested variant with ALE. If any improves the ALE score, apply it and repeat this step.
Polishing with only Illumina reads:
unicycler_polish -1 short_reads_1.fastq.gz -2 short_reads_2.fastq.gz -a assembly.fasta
Polishing with only PacBio reads:
unicycler_polish --pb_bax path/to/*bax.h5 -a assembly.fasta
Hybrid read set (Illumina and PacBio) polishing:
unicycler_polish -1 short_reads_1.fastq.gz -2 short_reads_2.fastq.gz --pb_bax *bax.h5 -a assembly.fasta
Hybrid read set (Illumina and Nanopore) polishing:
unicycler_polish -1 short_reads_1.fastq.gz -2 short_reads_2.fastq.gz --long_reads nanopore.fastq.gz -a assembly.fasta
A preprint version of the Unicycler manuscript is currently available on bioRxiv: biorxiv.org/content/early/2016/12/22/096412
Unicycler would not have been possible without Kat Holt, my fellow researchers in her lab and the many other people I work with at the University of Melbourne's Centre for Systems Genomics. In particular, Margaret Lam, Kelly Wyres and David Edwards worked with me on many challenging genomes during Unicycler's development.
Unicycler uses SeqAn to perform alignments and other sequence manipulations. The authors of this library have been very helpful during Unicycler's development and I owe them a great deal of thanks! It also uses minimap to seed alignments and so I'd like to thank Heng Li for writing such a fast long read aligner.