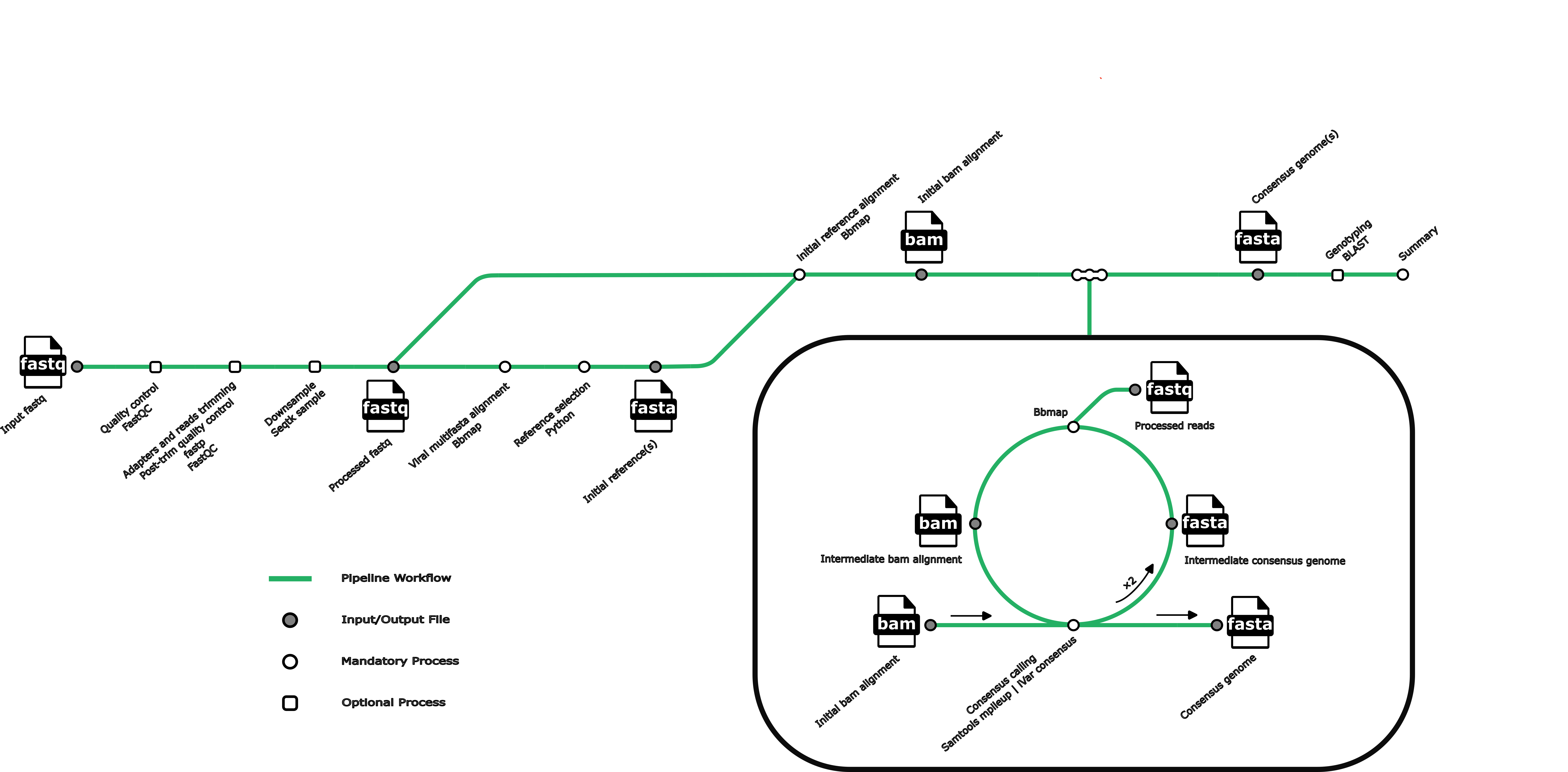
Revica is a reference-based viral consensus genome assembly pipeline for some of the most common respiratory viruses. Revica currently supports genome assembly of:
- Enterovirus (EV)
- Seasonal human coronavirus (HCOV)
- Human metapneumovirus (HMPV)
- Human respiratory syncytial virus (HRSV)
- Human parainfluenza virus (HPIV)
- Measles morbillivirus (MeV)
- Influenza A virus (IAV)
- Influenza B virus (IBV)
- Human adenovirus (HAdV)
Install Nextflow
Install Docker
To run Revica:
nextflow run asereewit/revica -r main -latest --input example_samplesheet.csv --output example_output -profile docker
on AWS:
nextflow run asereewit/revica -r main -latest --input example_samplesheet.csv --output example_output -profile docker -c your_nextflow_aws.config
| Option | Explanation |
|---|---|
--input |
samplesheet in csv format with fastq information |
--output |
output directory (default: revica_output) |
--db |
(multi)fasta file to overwrite the bundled viral database |
--run_name |
name for the summary tsv file (default: 'run') |
--skip_fastqc |
skip quality control using FastQC (default: false) |
--skip_fastp |
skip adapters and reads trimming using fastp (default: false) |
--run_kraken2 |
run Kraken2 for classifying reads (default: false) |
--kraken2_db |
Kraken2 database for reads classification, needs to be specified when using --run_kraken2 |
--kraken2_variants_host_filter |
use reads that didn't map to the kraken2 database for downstream consensus calling |
--save_kraken2_unclassified_reads |
save reads that didn't map to the specified kraken2 database |
--save_kraken2_classified_reads |
save reads that map to the specified kraken2 database |
--trim_len |
minimum read length to keep (default:50) |
--save_trimmed_reads |
save trimmed fastq |
--sample |
downsample fastq to a certain fraction or number of reads |
--ref_min_median_cov |
minimum median coverage on a reference for consensus assembly (default: 3) |
--ref_min_genome_cov |
minimum reference coverage percentage for consensus assembly (default: 60%) |
--ivar_consensus_t |
minimum frequency threshold to call consensus (default: 0.6) |
--ivar_consensus_q |
minimum quality score threshold to call consensus (default: 20) |
--ivar_consensus_m |
minimum depth to call consensus (default: 5) |
- Samplesheet example:
assets/samplesheet.csv - You can create a samplesheet using the bundled python script:
python bin/fastq_dir_samplesheet.py fastq_dir samplesheet_name.csv - Memory and CPU usage for pipeline processes can be adjusted in
conf/base.config - Process arguments can be adjusted in
conf/modules.config - You can use your own reference(s) for consensus genome assembly by specifying the
--dbparameter followed by your fasta file.- reference header format:
>reference_accession reference_tag reference_header_info - it's important to tag the fasta sequences for the same species or gene segments with the same name or abbreviation in the header section, otherwise the pipeline will generate a consensus genome for every reference where the median coverage of the first alignment exceed the specified threshold (default 3).
- Revica works with segmented viral genomes, just keep the different gene segments separated and tag them in the reference fasta file
- reference header format:
- If you are using Docker on Linux, check out these post-installation steps (especially cgroup swap limit capabilities support) for configuring Linux to work better with Docker.
- By default, Docker has full access to full RAM and CPU resources of the host, but if you are using MacOS, go to Settings -> Resources in Docker Desktop to make sure enough resources are allocated to docker containers.
For bug reports please email [email protected] or raise an issue on Github.