Bayesian genotyper for structural variants
svtyper \
-i sv.vcf \
-B sample.bam \
-l sample.bam.json \
> sv.gt.vcf
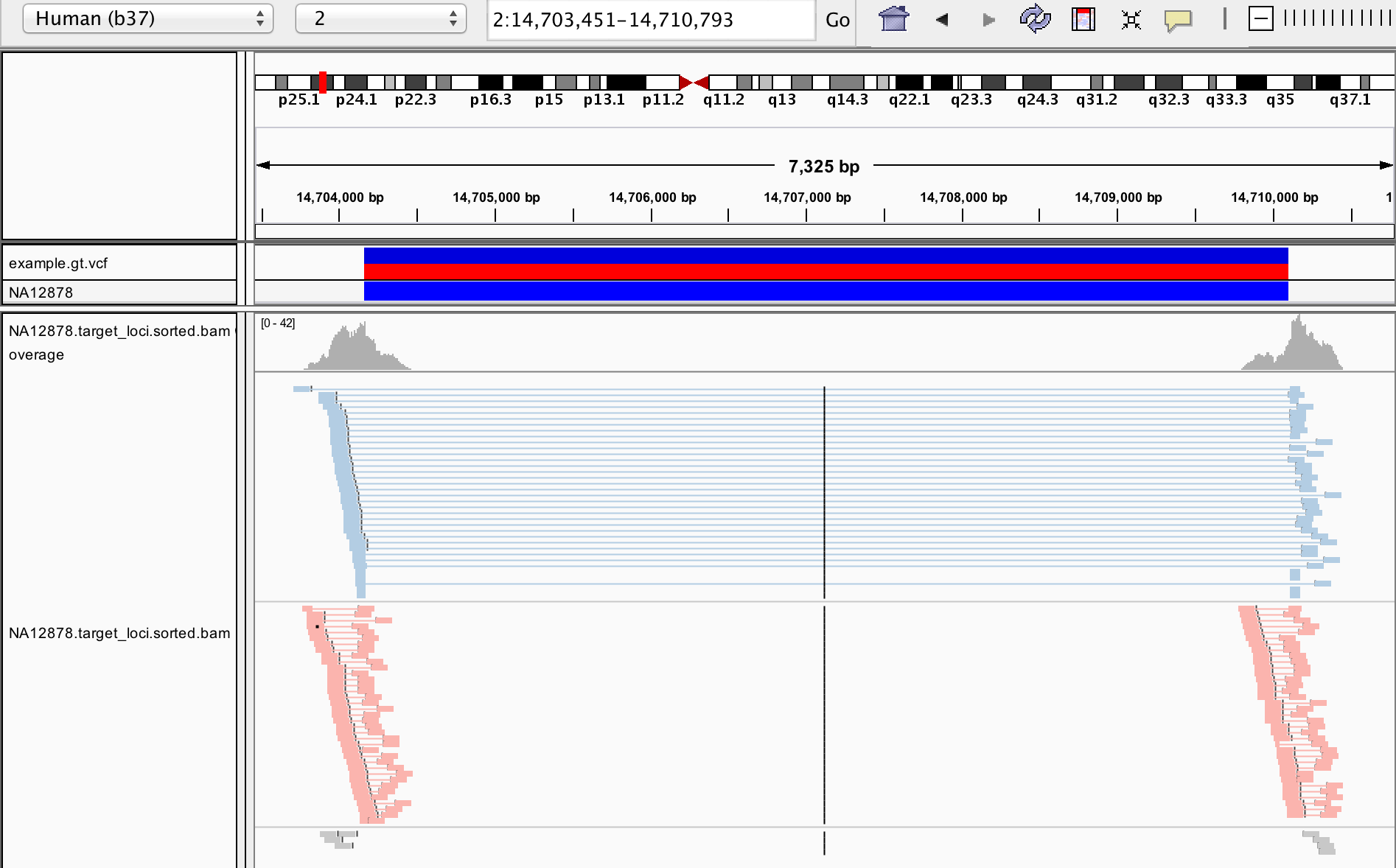
SVTyper performs breakpoint genotyping of structural variants (SVs) using whole genome sequencing data. Users must supply a VCF file of sites to genotype (which may be generated by LUMPY) as well as a BAM/CRAM file of Illumina paired-end reads aligned with BWA-MEM. SVTyper assesses discordant and concordant reads from paired-end and split-read alignments to infer genotypes at each site. Algorithm details and benchmarking are described in Chiang et al., 2015.
Requirements
- Python 2.7 or newer
- Pysam 0.8.1 or newer
Clone the repository
git clone [email protected]:hall-lab/svtyper.git
Test the installation
cd svtyper/test
../svtyper \
-i example.vcf \
-B NA12878.target_loci.sorted.bam \
-l NA12878.bam.json
> test.vcf
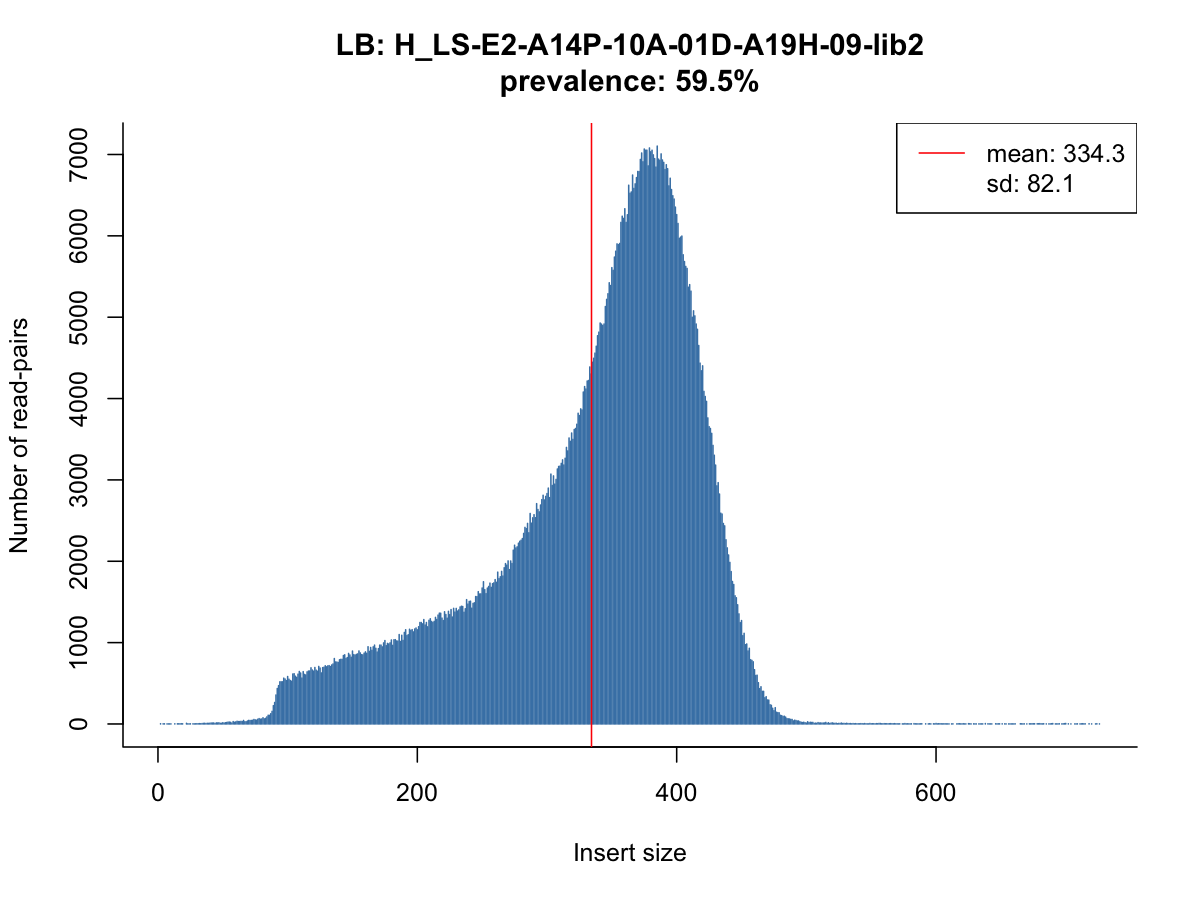
Many common issues are related to abnormal insert size distributions in the BAM file. SVTyper provides methods to assess and visualize the characteristics of sequencing libraries.
Running SVTyper with the -l flag creates a JSON file with essential metrics on a BAM file. SVTyper will sample the first N reads for the file (1 million by default) to parse the libraries, read groups, and insert size histograms. This can be done in the absence of a VCF file.
svtyper \
-B my.bam \
-l my.bam.json
The lib_stats.R script produces insert size histograms from the JSON file
scripts/lib_stats.R my.bam.json my.bam.json.pdf
C Chiang, R M Layer, G G Faust, M R Lindberg, D B Rose, E P Garrison, G T Marth, A R Quinlan, and I M Hall. SpeedSeq: ultra-fast personal genome analysis and interpretation. Nat Meth 12, 966–968 (2015). doi:10.1038/nmeth.3505.
http://www.nature.com/nmeth/journal/vaop/ncurrent/full/nmeth.3505.html